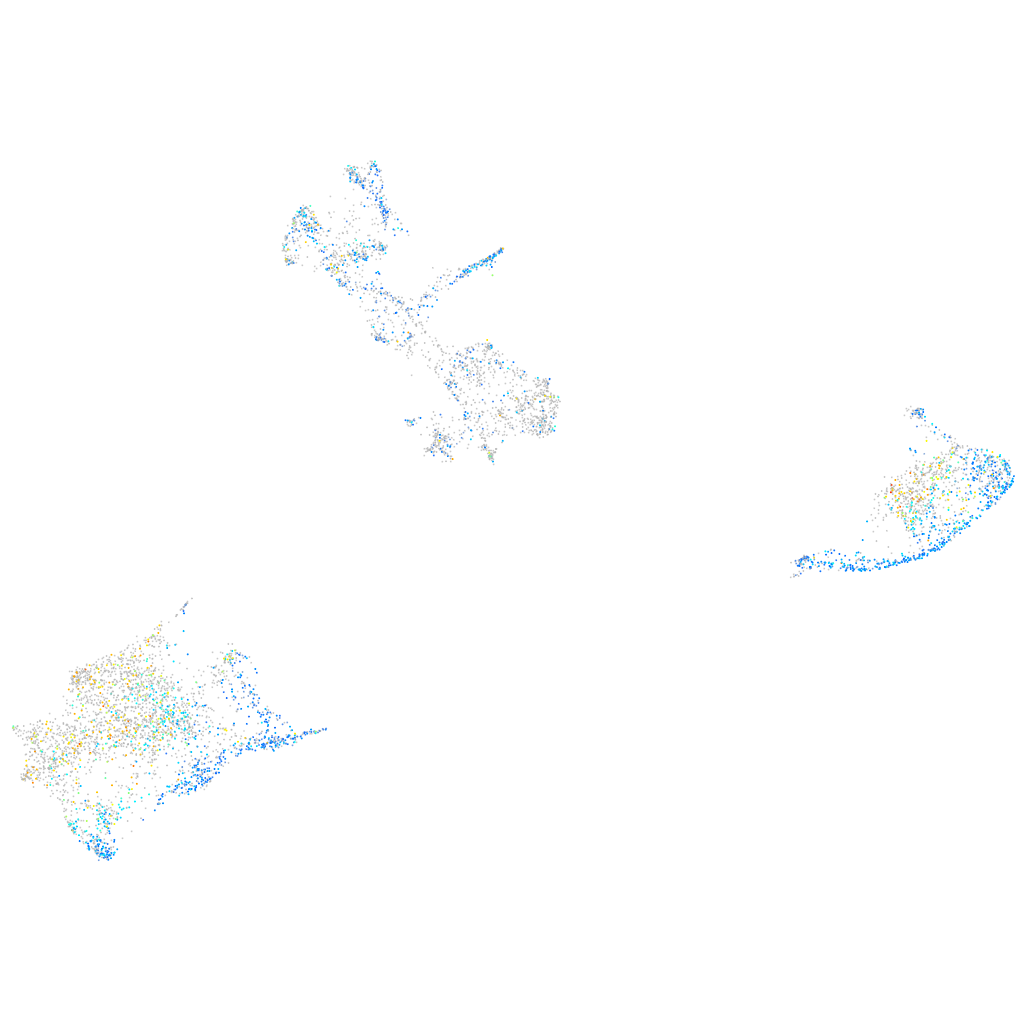
si:ch211-108c6.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rab38 | 0.180 | si:ch211-222l21.1 | -0.129 |
| tspan36 | 0.167 | ptmaa | -0.125 |
| slc37a2 | 0.164 | si:ch73-1a9.3 | -0.122 |
| sptbn1 | 0.162 | hmgb1b | -0.110 |
| coro1ca | 0.160 | mdkb | -0.108 |
| zgc:91968 | 0.156 | si:ch73-281n10.2 | -0.108 |
| tspan10 | 0.154 | gpm6aa | -0.108 |
| bace2 | 0.153 | h3f3a | -0.108 |
| tmem243a | 0.152 | stmn1a | -0.107 |
| qdpra | 0.150 | nova2 | -0.107 |
| syngr1a | 0.147 | hmgb3a | -0.105 |
| ahnak | 0.147 | si:ch211-288g17.3 | -0.104 |
| mlpha | 0.146 | hmgb2a | -0.102 |
| rabl6b | 0.144 | hmgn2 | -0.102 |
| SPAG9 | 0.144 | hmgb2b | -0.100 |
| gpr143 | 0.143 | marcksl1a | -0.099 |
| slc24a5 | 0.142 | ptmab | -0.098 |
| tyr | 0.142 | stmn1b | -0.096 |
| mitfa | 0.141 | elavl3 | -0.095 |
| cdh1 | 0.140 | tmsb | -0.095 |
| cdk15 | 0.140 | epb41a | -0.094 |
| pttg1ipb | 0.139 | tuba1c | -0.094 |
| si:zfos-943e10.1 | 0.137 | gng3 | -0.094 |
| si:ch73-389b16.1 | 0.135 | sox11a | -0.094 |
| C12orf75 | 0.135 | zc4h2 | -0.089 |
| abracl | 0.135 | sox11b | -0.089 |
| pmela | 0.132 | CU467822.1 | -0.087 |
| rab34b | 0.132 | scrt2 | -0.085 |
| lamp1a | 0.131 | sncb | -0.083 |
| tfap2e | 0.130 | tubb2b | -0.083 |
| slc3a2a | 0.130 | cadm3 | -0.081 |
| dct | 0.129 | zgc:100920 | -0.077 |
| pcbd1 | 0.129 | olfm1b | -0.076 |
| tyrp1b | 0.125 | pcna | -0.076 |
| ap3d1 | 0.125 | nat8l | -0.076 |