si:ch1073-153i20.4
ZFIN 
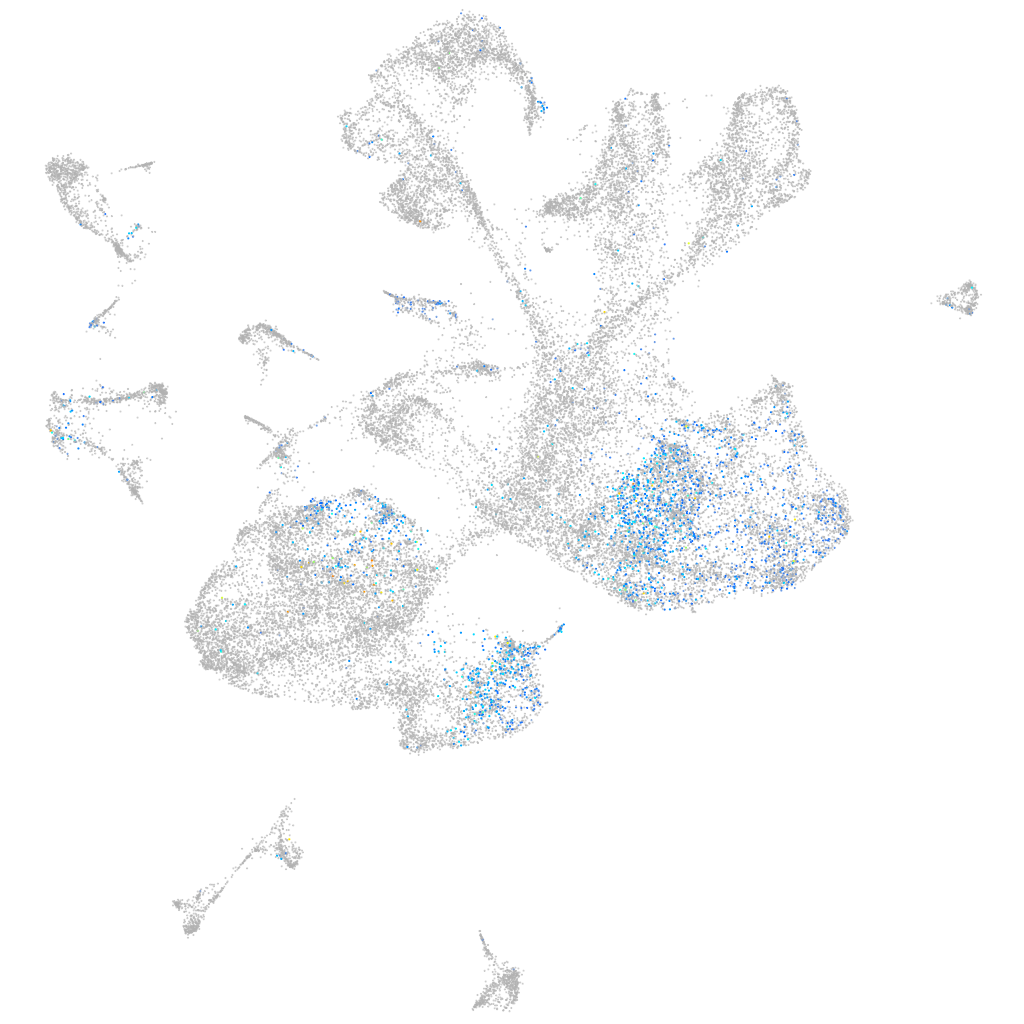
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110216 | 0.351 | ckbb | -0.134 |
| zgc:165555.3 | 0.341 | rtn1a | -0.122 |
| si:dkey-261m9.17 | 0.322 | gpm6ab | -0.105 |
| mibp | 0.314 | ppdpfb | -0.100 |
| si:ch211-113a14.18 | 0.314 | si:dkeyp-75h12.5 | -0.099 |
| si:dkey-108k21.10 | 0.312 | rnasekb | -0.099 |
| zgc:110425 | 0.307 | elavl3 | -0.095 |
| fbxo5 | 0.295 | gng3 | -0.095 |
| hist1h4l | 0.289 | gapdhs | -0.094 |
| hist1h2a11 | 0.286 | calm1a | -0.093 |
| mki67 | 0.286 | fez1 | -0.092 |
| ccna2 | 0.277 | atp6v1e1b | -0.092 |
| si:ch211-113a14.24 | 0.275 | elavl4 | -0.089 |
| cks1b | 0.274 | sncb | -0.089 |
| zgc:153405 | 0.266 | cotl1 | -0.089 |
| lbr | 0.265 | atp6v0cb | -0.088 |
| smc2 | 0.262 | myt1b | -0.088 |
| aurkb | 0.258 | stmn1b | -0.088 |
| ncapg | 0.254 | stxbp1a | -0.087 |
| dek | 0.253 | pvalb1 | -0.087 |
| rrm2 | 0.252 | pvalb2 | -0.086 |
| top2a | 0.243 | gabarapb | -0.086 |
| mad2l1 | 0.243 | vamp2 | -0.085 |
| rrm1 | 0.241 | atp6v1g1 | -0.085 |
| cdk1 | 0.240 | snap25a | -0.085 |
| tuba8l | 0.236 | acbd7 | -0.083 |
| dut | 0.235 | slc1a2b | -0.083 |
| tpx2 | 0.231 | cadm4 | -0.082 |
| chaf1a | 0.228 | rtn1b | -0.082 |
| cdca5 | 0.226 | dpysl3 | -0.081 |
| hmgb2a | 0.220 | tmsb | -0.081 |
| banf1 | 0.220 | mllt11 | -0.081 |
| ncapd2 | 0.217 | zc4h2 | -0.081 |
| stmn1a | 0.217 | stx1b | -0.080 |
| kifc1 | 0.216 | gabarapl2 | -0.079 |