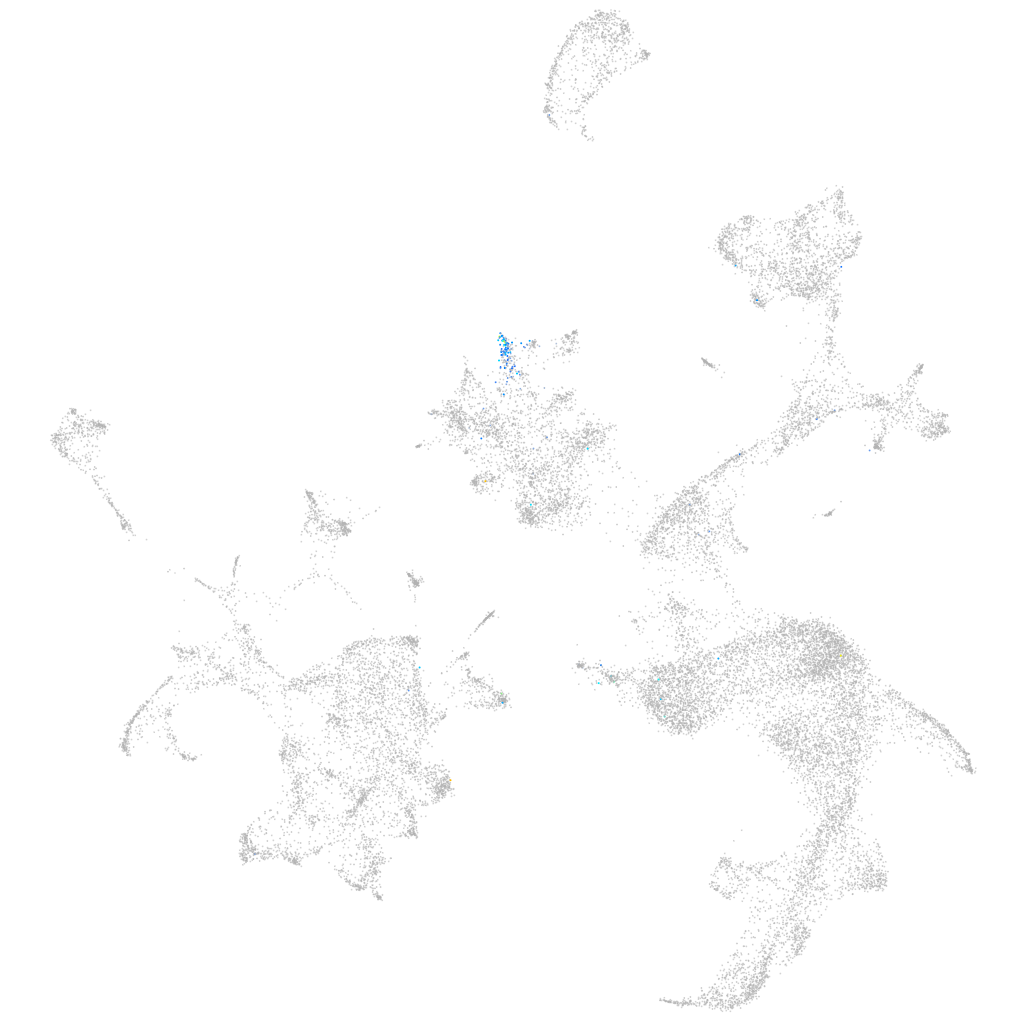
si:cabz01076231.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye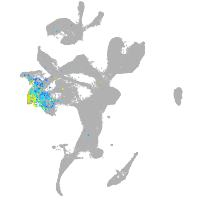 |
taste / olfactory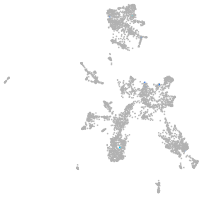 |
otic / lateral line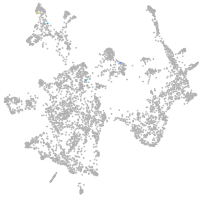 |
epidermis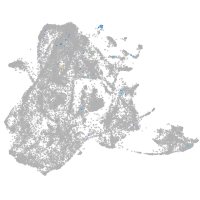 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prph2a | 0.443 | hbbe3 | -0.032 |
| rcvrn3 | 0.441 | msna | -0.030 |
| gnat2 | 0.436 | mob1a | -0.027 |
| inaa | 0.421 | she | -0.023 |
| prph2b | 0.412 | etf1b | -0.023 |
| pde6h | 0.408 | f13a1b | -0.022 |
| gnb3b | 0.403 | vamp3 | -0.022 |
| pde6c | 0.400 | pdap1a | -0.022 |
| si:ch1073-469d17.2 | 0.398 | egfl7 | -0.022 |
| zgc:73359 | 0.396 | lmo2 | -0.022 |
| grk1b | 0.396 | anxa11a | -0.021 |
| rbp4l | 0.394 | gng5 | -0.021 |
| guca1c | 0.391 | si:ch211-114n24.6 | -0.021 |
| pde6ha | 0.389 | akap12b | -0.021 |
| si:ch211-81a5.8 | 0.388 | tram1 | -0.020 |
| cngb3.2 | 0.387 | snx1a | -0.020 |
| pdcb | 0.384 | ptbp1a | -0.020 |
| rcvrn2 | 0.383 | srgn | -0.020 |
| arr3a | 0.382 | urod | -0.020 |
| CT573282.1 | 0.378 | rhocb | -0.019 |
| grk7a | 0.371 | clec14a | -0.019 |
| syt5a | 0.370 | rnaseka | -0.019 |
| slc25a3a | 0.368 | clic2 | -0.019 |
| ckmt2a | 0.360 | sept2 | -0.019 |
| cnga3a | 0.357 | zgc:56493 | -0.019 |
| arl3l2 | 0.353 | cdh5 | -0.019 |
| si:ch1073-303d10.1 | 0.352 | arl4ab | -0.019 |
| guk1b | 0.347 | magt1 | -0.019 |
| opn1lw2 | 0.339 | pecam1 | -0.018 |
| tmem237a | 0.337 | blvra | -0.018 |
| zgc:195245 | 0.334 | kdrl | -0.018 |
| rgs9a | 0.328 | aldoaa | -0.018 |
| aqp9b | 0.327 | podxl | -0.018 |
| rp1 | 0.322 | tagln2 | -0.018 |
| si:dkey-17e16.15 | 0.314 | btk | -0.018 |