si:cabz01054394.7
ZFIN 
Other cell groups


all cells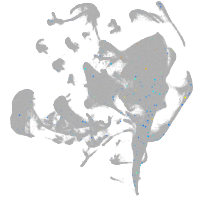 |
blastomeres |
PGCs |
endoderm |
axial mesoderm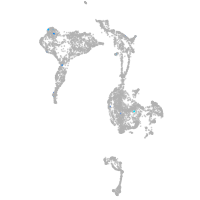 |
muscle 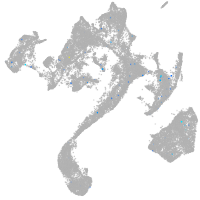 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 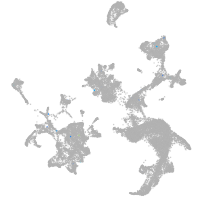 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis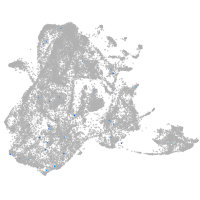 |
periderm |
fin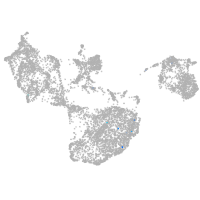 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.026 | pvalb1 | -0.014 |
| dkc1 | 0.025 | elavl3 | -0.012 |
| nhp2 | 0.025 | hbae3 | -0.012 |
| nop56 | 0.025 | pvalb2 | -0.012 |
| nop58 | 0.025 | actc1b | -0.011 |
| snu13b | 0.025 | gpm6aa | -0.010 |
| fbl | 0.024 | hbae1.1 | -0.010 |
| mki67 | 0.024 | hbbe1.3 | -0.010 |
| anp32b | 0.023 | stmn1b | -0.010 |
| cnbpa | 0.023 | tmsb | -0.010 |
| nop10 | 0.023 | ckbb | -0.009 |
| nop2 | 0.023 | gpm6ab | -0.009 |
| ptges3b | 0.023 | hbbe1.1 | -0.009 |
| ranbp1 | 0.023 | mylpfa | -0.009 |
| banf1 | 0.022 | rtn1a | -0.009 |
| eif5a2 | 0.022 | adgrl3.1 | -0.008 |
| emg1 | 0.022 | gng3 | -0.008 |
| gnl3 | 0.022 | hbae1.3 | -0.008 |
| hspb1 | 0.022 | myt1b | -0.008 |
| mad2l1 | 0.022 | myhz1.1 | -0.008 |
| ncl | 0.022 | clvs2 | -0.007 |
| nutf2l | 0.022 | hbbe2 | -0.007 |
| pa2g4a | 0.022 | mllt11 | -0.007 |
| rrp1 | 0.022 | mylz3 | -0.007 |
| tpx2 | 0.022 | nova2 | -0.007 |
| wdr43 | 0.022 | nsg2 | -0.007 |
| BX927258.1 | 0.021 | pbx1a | -0.007 |
| bxdc2 | 0.021 | rnasekb | -0.007 |
| cad | 0.021 | scrt2 | -0.007 |
| cbx3a | 0.021 | sox4a | -0.007 |
| cdca8 | 0.021 | tnni2a.4 | -0.007 |
| ebna1bp2 | 0.021 | tpma | -0.007 |
| lyar | 0.021 | tuba1c | -0.007 |
| nip7 | 0.021 | calm1a | -0.007 |
| rpl7l1 | 0.021 | adam22 | -0.006 |




