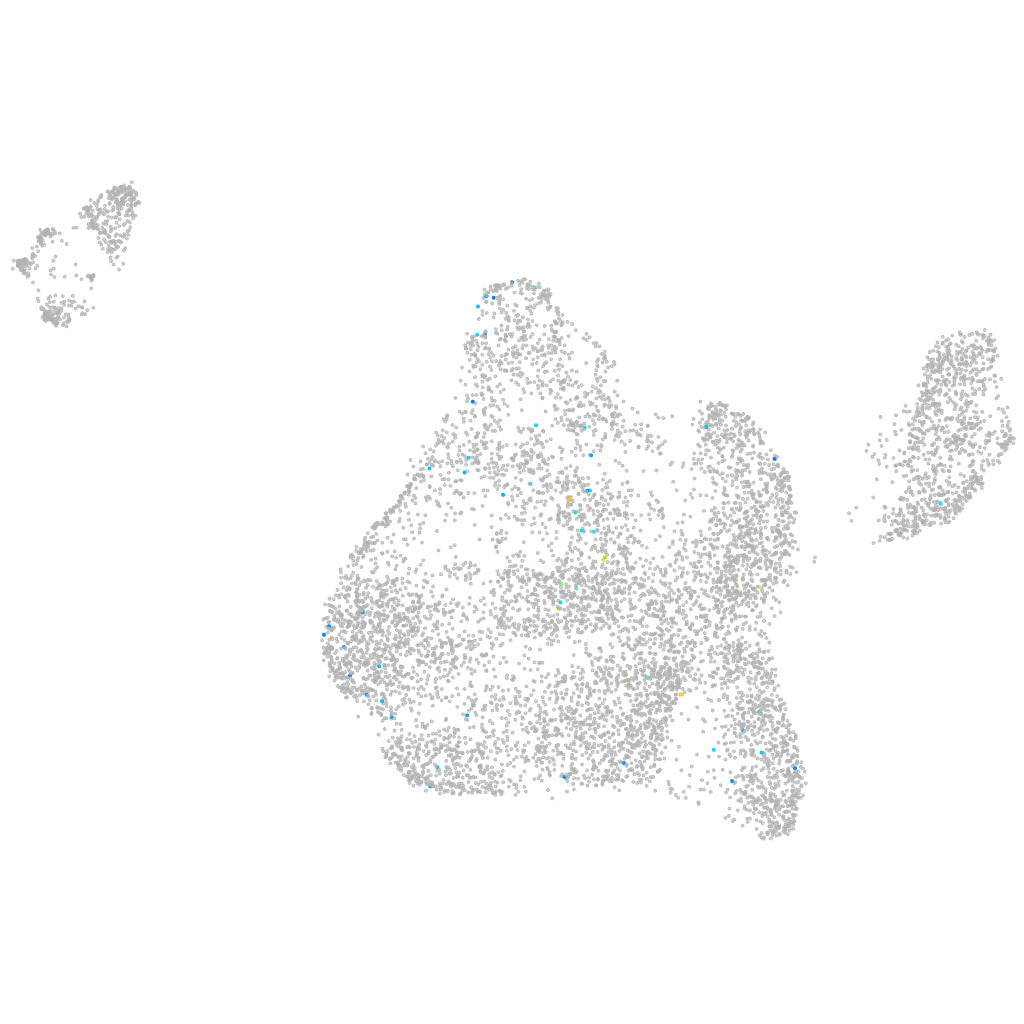
si:busm1-194e12.11
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lingo3a | 0.169 | zgc:173742 | -0.042 |
| CABZ01081897.1 | 0.165 | h1m | -0.039 |
| si:ch211-285j22.5 | 0.161 | thy1 | -0.039 |
| CR931802.2 | 0.158 | acp5a | -0.038 |
| ponzr4 | 0.156 | pttg1 | -0.038 |
| BX537308.1 | 0.139 | epcam | -0.037 |
| camk1gb | 0.131 | blf | -0.037 |
| LOC103910957 | 0.130 | rbm7 | -0.037 |
| FO904898.6 | 0.130 | cldnd | -0.037 |
| LOC103909736 | 0.130 | prmt1 | -0.035 |
| XLOC-033352 | 0.124 | spink2.1 | -0.035 |
| si:dkey-219e21.4 | 0.124 | gstp1 | -0.035 |
| BX323812.2 | 0.122 | npc2 | -0.034 |
| dnase1l1l | 0.122 | tmem165 | -0.034 |
| v2rh1 | 0.118 | nsmce4a | -0.034 |
| CU571315.2 | 0.114 | glulb | -0.033 |
| FO818743.1 | 0.113 | ccna1 | -0.033 |
| olfml3b | 0.113 | psme3 | -0.033 |
| ca6 | 0.109 | ska2 | -0.033 |
| ccr2 | 0.105 | tcf25 | -0.033 |
| fat1b | 0.105 | drap1 | -0.033 |
| spon2b | 0.104 | zgc:100951 | -0.032 |
| axdnd1 | 0.103 | pus1 | -0.032 |
| cldn7a | 0.103 | si:ch211-132b12.8 | -0.032 |
| gpm6aa | 0.102 | FO834825.1 | -0.032 |
| XLOC-001835 | 0.101 | h2af1al | -0.031 |
| cfap157 | 0.098 | si:ch73-299h12.3 | -0.031 |
| XLOC-043980 | 0.097 | stk35l | -0.030 |
| BX664625.2 | 0.096 | hyou1 | -0.030 |
| gabrp | 0.093 | fntb | -0.029 |
| ihha | 0.093 | zcchc7 | -0.029 |
| prob1 | 0.092 | aktip | -0.029 |
| cdk5r1b | 0.091 | cox17 | -0.029 |
| AL954861.2 | 0.091 | ppp1r2 | -0.029 |
| LOC110439513 | 0.091 | siva1 | -0.028 |