SHC (Src homology 2 domain containing) transforming protein 2
ZFIN 
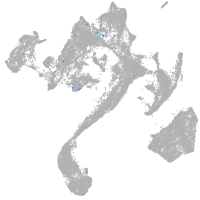
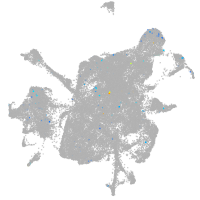
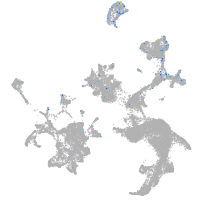
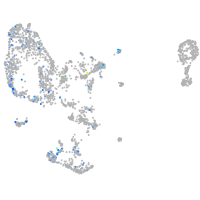
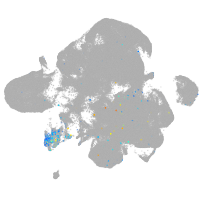
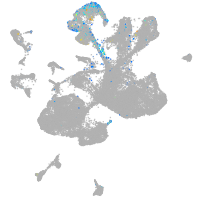
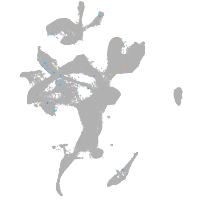
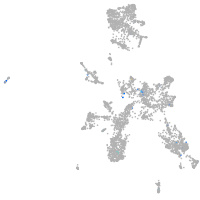
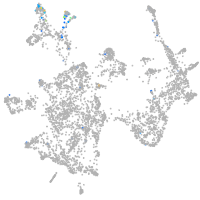
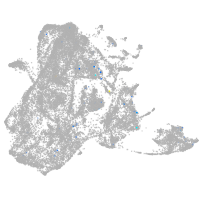
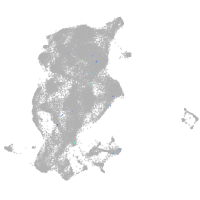
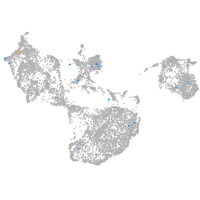
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| isl1 | 0.266 | id1 | -0.135 |
| isl2a | 0.256 | XLOC-003690 | -0.133 |
| onecut1 | 0.246 | mdka | -0.123 |
| chodl | 0.236 | cldn5a | -0.115 |
| slit3 | 0.216 | sox3 | -0.113 |
| slc18a3a | 0.204 | her15.1 | -0.112 |
| prph | 0.190 | notch3 | -0.107 |
| gfra3 | 0.189 | XLOC-003689 | -0.099 |
| phox2a | 0.187 | sparc | -0.098 |
| ret | 0.182 | sdc4 | -0.095 |
| phox2bb | 0.182 | btg2 | -0.093 |
| tbx20 | 0.181 | sox19a | -0.092 |
| cxcr4b | 0.179 | cd82a | -0.091 |
| islr2 | 0.178 | her4.1 | -0.090 |
| si:ch211-247j9.1 | 0.174 | her12 | -0.088 |
| onecutl | 0.170 | COX7A2 (1 of many) | -0.088 |
| gap43 | 0.168 | sox2 | -0.086 |
| elavl3 | 0.168 | CU467822.1 | -0.086 |
| tmsb | 0.165 | vamp3 | -0.085 |
| pacsin1b | 0.165 | sb:cb81 | -0.085 |
| onecut2 | 0.165 | tspan7 | -0.085 |
| nrp1a | 0.163 | gpm6bb | -0.085 |
| gng2 | 0.163 | atp1a1b | -0.085 |
| gab2 | 0.161 | fabp7a | -0.084 |
| stmn1b | 0.161 | XLOC-003692 | -0.083 |
| lmo4b | 0.156 | hmgb2a | -0.083 |
| adcyap1b | 0.156 | msi1 | -0.083 |
| gng3 | 0.156 | gfap | -0.082 |
| agrn | 0.152 | si:ch211-286b5.5 | -0.081 |
| ebf1a | 0.152 | fosab | -0.081 |
| map1b | 0.152 | pgrmc1 | -0.080 |
| rtn1a | 0.151 | her4.2 | -0.080 |
| nefmb | 0.150 | gas1b | -0.079 |
| lhx4 | 0.150 | zgc:165461 | -0.079 |
| XLOC-007078 | 0.149 | notch1b | -0.076 |