SFT2 domain containing 1
ZFIN 
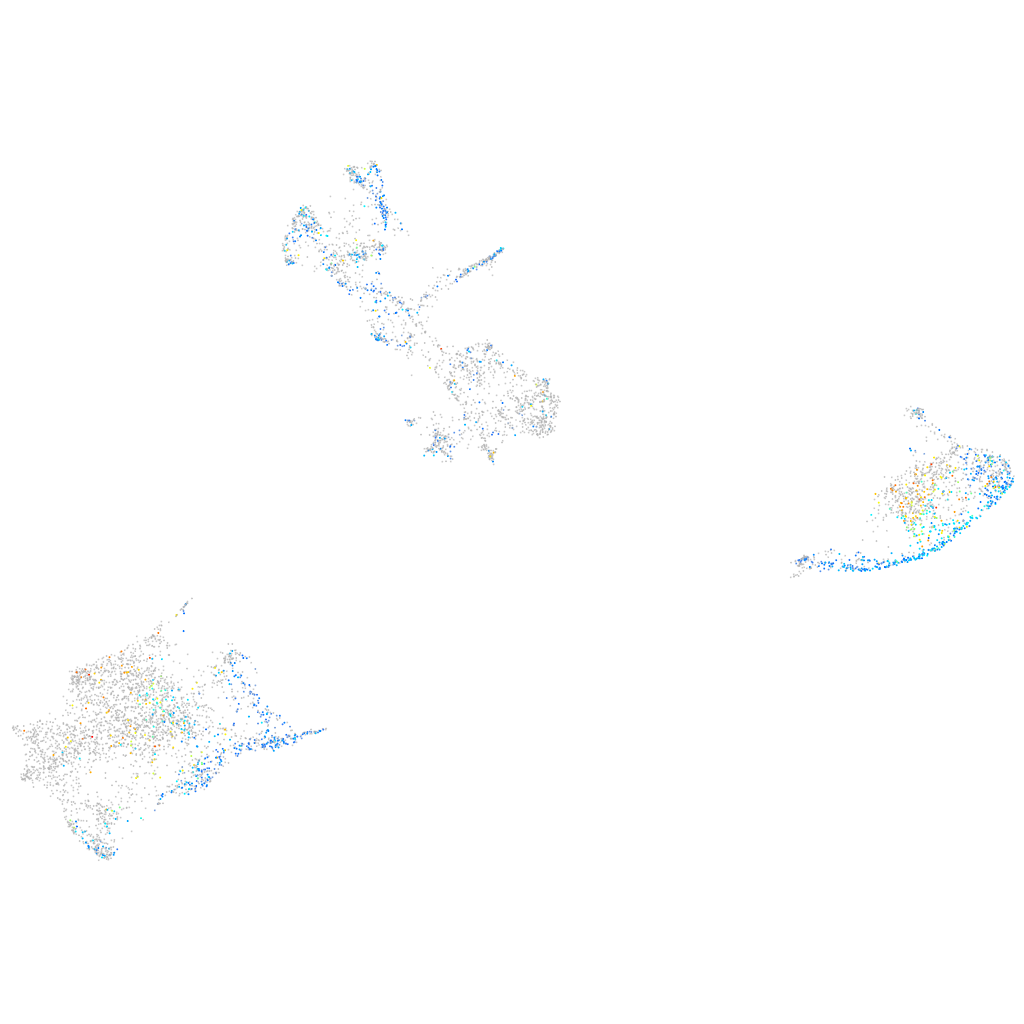
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.277 | CABZ01021592.1 | -0.134 |
| slc24a5 | 0.276 | pvalb2 | -0.098 |
| tyrp1a | 0.265 | ptmaa | -0.098 |
| slc37a2 | 0.254 | pvalb1 | -0.096 |
| pmela | 0.253 | si:dkey-251i10.2 | -0.094 |
| tyr | 0.252 | uraha | -0.093 |
| oca2 | 0.252 | si:ch211-222l21.1 | -0.093 |
| atp6v0ca | 0.252 | gpm6aa | -0.087 |
| slc45a2 | 0.251 | stmn1b | -0.087 |
| tyrp1b | 0.249 | apoa2 | -0.086 |
| tspan36 | 0.242 | tmsb | -0.085 |
| zgc:91968 | 0.241 | si:ch73-1a9.3 | -0.085 |
| qdpra | 0.239 | hmgb1b | -0.085 |
| lamp1a | 0.233 | elavl3 | -0.084 |
| tspan10 | 0.226 | CU467822.1 | -0.082 |
| si:ch73-389b16.1 | 0.226 | nova2 | -0.082 |
| slc29a3 | 0.225 | tuba1c | -0.079 |
| aadac | 0.219 | gng3 | -0.077 |
| SPAG9 | 0.217 | hmgb2b | -0.076 |
| atp6v0a2b | 0.216 | epb41a | -0.075 |
| slc3a2a | 0.215 | actc1b | -0.074 |
| slc7a5 | 0.215 | zc4h2 | -0.074 |
| pah | 0.215 | sncb | -0.073 |
| kita | 0.214 | stmn1a | -0.071 |
| gpr143 | 0.209 | si:ch211-251b21.1 | -0.071 |
| rab38 | 0.207 | cdh2 | -0.069 |
| rab32a | 0.206 | marcksl1a | -0.069 |
| atp6ap1b | 0.205 | hmgn2 | -0.069 |
| prkar1b | 0.201 | dio3a | -0.069 |
| mtbl | 0.201 | LOC110437731 | -0.068 |
| naga | 0.200 | foxp4 | -0.068 |
| syngr1a | 0.199 | gng2 | -0.067 |
| AL935199.1 | 0.198 | aqp3a | -0.067 |
| atp11a | 0.197 | mdkb | -0.067 |
| bace2 | 0.196 | ppdpfb | -0.066 |