"serpin peptidase inhibitor, clade G (C1 inhibitor), member 1"
ZFIN 
Other cell groups


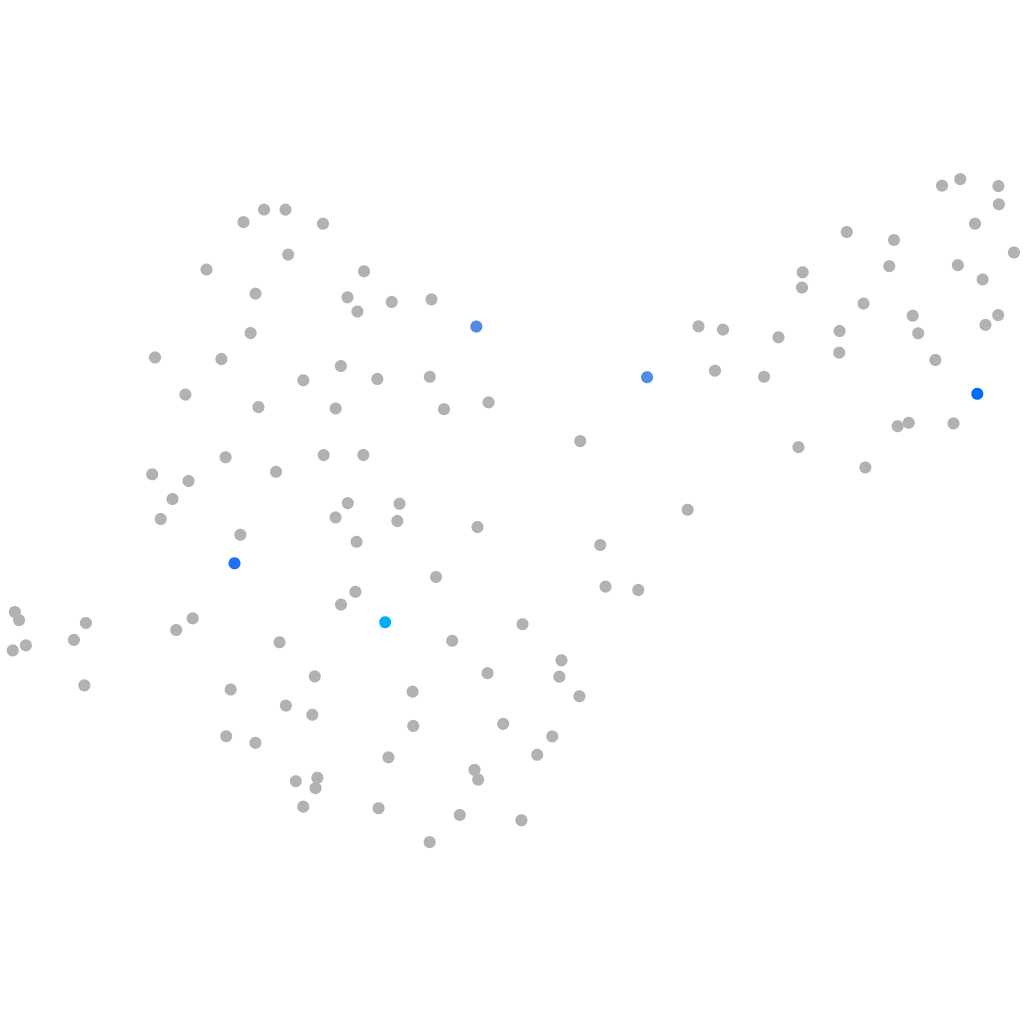
all cells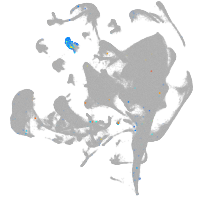 |
blastomeres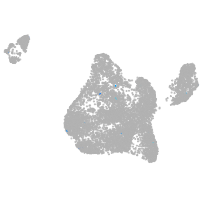 |
PGCs |
endoderm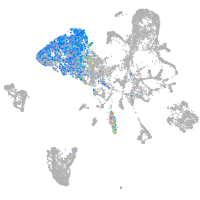 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX649516.2 | 0.712 | ctsba | -0.206 |
| fancb | 0.712 | sec62 | -0.200 |
| gmip | 0.712 | elavl1a | -0.192 |
| plk4 | 0.712 | cxcr4b | -0.192 |
| sox32 | 0.712 | zgc:77118 | -0.190 |
| abhd15a | 0.627 | dkc1 | -0.186 |
| unc5da | 0.618 | kat7b | -0.183 |
| si:ch1073-170o4.1 | 0.585 | srsf11 | -0.183 |
| exorh | 0.584 | sf3b1 | -0.176 |
| unc45a | 0.580 | ddit4 | -0.173 |
| twist1a | 0.579 | nono | -0.165 |
| chrnb2l | 0.578 | cnbpa | -0.165 |
| pik3r5 | 0.578 | ccnb1 | -0.163 |
| zap70 | 0.578 | c1d | -0.157 |
| si:ch211-12h2.6 | 0.573 | lrrc59 | -0.155 |
| GALNT3 | 0.568 | rtf1 | -0.155 |
| s1pr2 | 0.563 | epcam | -0.152 |
| hey1 | 0.550 | CT030188.1 | -0.150 |
| CT025748.1 | 0.540 | pa2g4b | -0.150 |
| CU855889.2 | 0.523 | kif11 | -0.149 |
| ca2 | 0.522 | cenpf | -0.149 |
| slc35a5 | 0.517 | tmed9 | -0.148 |
| pebp1 | 0.516 | si:ch211-262h13.3 | -0.147 |
| met | 0.516 | gnl3 | -0.147 |
| adgrg11 | 0.510 | ddx61 | -0.144 |
| adora2aa | 0.510 | nup58 | -0.142 |
| jag1a | 0.510 | si:ch73-347e22.4 | -0.142 |
| thbs4a | 0.510 | chtf18 | -0.141 |
| b9d2 | 0.509 | marcksa | -0.141 |
| pygmb | 0.494 | nol7 | -0.141 |
| trim37 | 0.488 | actb1 | -0.140 |
| clcn6 | 0.485 | kmt5ab | -0.140 |
| LOC101884641 | 0.476 | syncripl | -0.139 |
| efna1a | 0.476 | prelid3b | -0.139 |
| plin2 | 0.476 | uck2b | -0.137 |