septin 3
ZFIN 
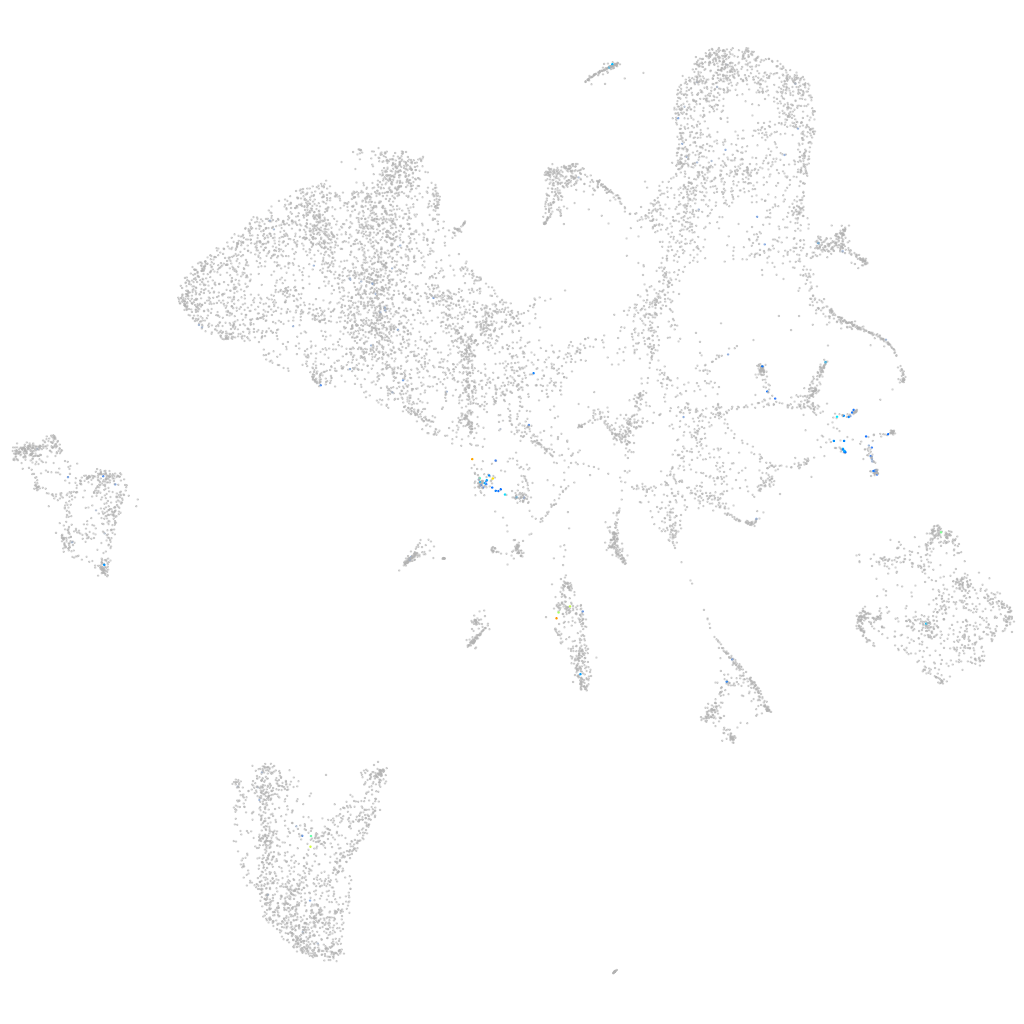
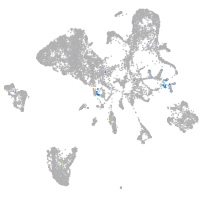
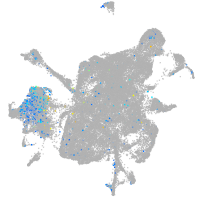
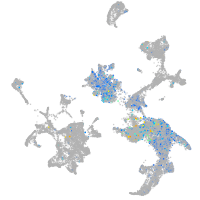
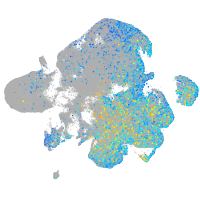
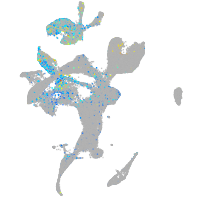
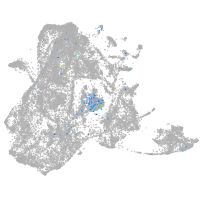
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fcer1g | 0.245 | aldob | -0.064 |
| myf6 | 0.234 | gstr | -0.063 |
| LOC101885646 | 0.228 | gapdh | -0.059 |
| adam22 | 0.224 | eno3 | -0.057 |
| PLPP7 (1 of many) | 0.217 | hdlbpa | -0.057 |
| si:ch211-278j3.3 | 0.216 | aldh6a1 | -0.057 |
| hoxc13b | 0.216 | ahcy | -0.055 |
| LOC100537384 | 0.213 | eef1da | -0.055 |
| AL935294.2 | 0.209 | hspe1 | -0.054 |
| tuba1c | 0.209 | cebpd | -0.054 |
| cacng8b | 0.208 | nme2b.1 | -0.053 |
| XLOC-025509 | 0.207 | sod1 | -0.053 |
| nsg2 | 0.207 | gstt1a | -0.051 |
| AL929222.2 | 0.204 | atp5l | -0.051 |
| rtn1b | 0.202 | cox6a1 | -0.050 |
| nlgn3b | 0.200 | ckba | -0.050 |
| elavl3 | 0.198 | gpx1a | -0.050 |
| gpc1a | 0.192 | prdx2 | -0.049 |
| nova2 | 0.190 | sdr16c5b | -0.049 |
| csdc2a | 0.188 | scp2a | -0.048 |
| lhx1b | 0.186 | glud1b | -0.048 |
| CU896691.1 | 0.186 | gcshb | -0.048 |
| tuba1a | 0.185 | agxta | -0.048 |
| mb21d2 | 0.185 | rps18 | -0.047 |
| akap6 | 0.184 | rpl6 | -0.047 |
| dmtn | 0.182 | abat | -0.047 |
| LOC100537369 | 0.180 | fbp1b | -0.047 |
| tmem91 | 0.179 | gamt | -0.046 |
| pknox2 | 0.178 | sod2 | -0.046 |
| BX890548.1 | 0.177 | cox8b | -0.046 |
| nhlh2 | 0.177 | mgst1.2 | -0.046 |
| XLOC-019975 | 0.175 | shmt1 | -0.046 |
| mthfd2l | 0.170 | nipsnap3a | -0.046 |
| uncx | 0.169 | gsta.1 | -0.046 |
| si:dkey-40m6.8 | 0.167 | suclg2 | -0.046 |