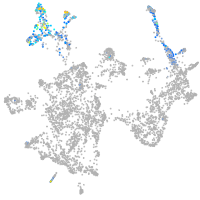
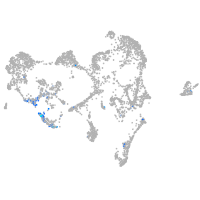
semaphorin 7A
ZFIN 
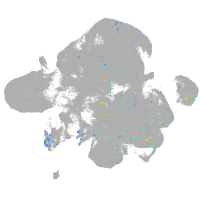
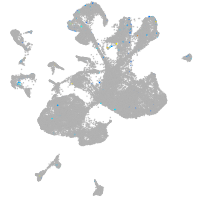
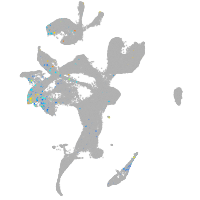
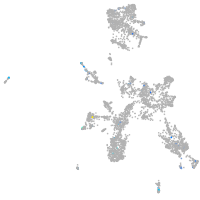
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fli1b | 0.333 | fabp3 | -0.070 |
| flt1 | 0.277 | aqp12 | -0.067 |
| adgrl4 | 0.256 | bhmt | -0.066 |
| LOC103909874 | 0.251 | agxtb | -0.066 |
| si:ch211-1a19.3 | 0.241 | apoc1 | -0.065 |
| LOC100332187 | 0.225 | tfa | -0.060 |
| LOC108179520 | 0.219 | kng1 | -0.059 |
| si:ch211-160d20.5 | 0.214 | cfhl4 | -0.059 |
| LOC108180727 | 0.211 | fetub | -0.058 |
| notchl | 0.209 | ces2 | -0.058 |
| slc2a11a | 0.209 | grhprb | -0.058 |
| myct1b | 0.206 | ambp | -0.058 |
| CR450686.1 | 0.205 | apoa2 | -0.058 |
| sc:d189 | 0.201 | apom | -0.058 |
| myct1a | 0.194 | hao1 | -0.057 |
| FO905052.1 | 0.193 | serpina1 | -0.057 |
| pecam1 | 0.191 | zgc:123103 | -0.057 |
| LOC101884756 | 0.186 | ttc36 | -0.057 |
| exoc3l1 | 0.185 | fgg | -0.056 |
| cdh5 | 0.185 | rbp2b | -0.056 |
| mmrn2a | 0.176 | fabp10a | -0.056 |
| scarf1 | 0.173 | ttr | -0.055 |
| sox18 | 0.169 | serpina1l | -0.055 |
| hlx1 | 0.169 | f2 | -0.055 |
| edn2 | 0.168 | c9 | -0.055 |
| XLOC-024725 | 0.167 | gc | -0.055 |
| slc4a11 | 0.166 | wu:fj16a03 | -0.055 |
| fgd5a | 0.162 | hpda | -0.055 |
| fam160a1b | 0.160 | uox | -0.054 |
| LOC101882124 | 0.152 | shbg | -0.053 |
| BX927369.1 | 0.144 | vtnb | -0.053 |
| ecscr | 0.144 | fgb | -0.053 |
| prokr1b | 0.144 | fga | -0.053 |
| erg | 0.141 | LOC110437731 | -0.052 |
| mmp16b | 0.141 | c3a.1 | -0.052 |