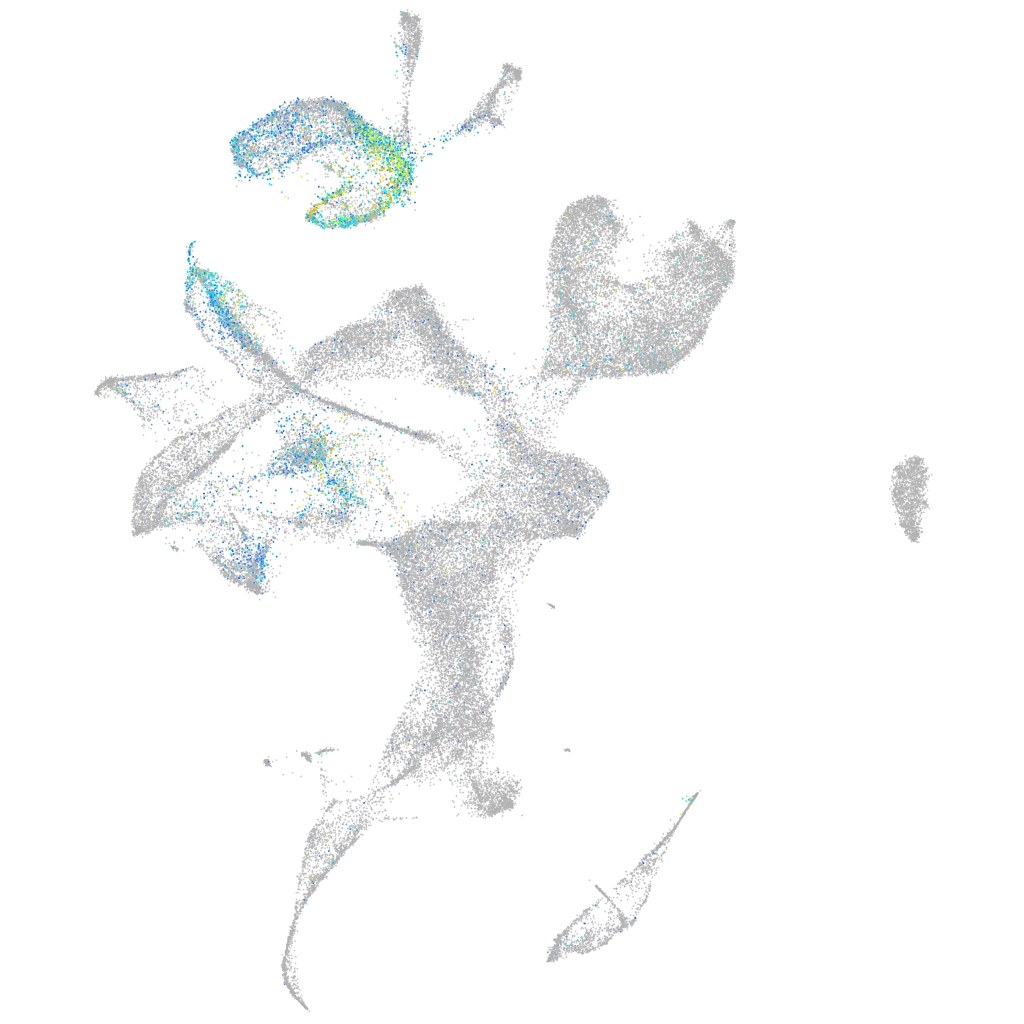scratch family zinc finger 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| s1pr1 | 0.295 | hmgb2a | -0.164 |
| elavl3 | 0.272 | pcna | -0.120 |
| barhl2 | 0.247 | msna | -0.114 |
| pax6b | 0.239 | selenoh | -0.112 |
| meis2b | 0.234 | mki67 | -0.111 |
| mab21l1 | 0.234 | chaf1a | -0.109 |
| prdm13 | 0.232 | dek | -0.108 |
| pax6a | 0.226 | rrm1 | -0.108 |
| mir181b-3 | 0.221 | ccna2 | -0.108 |
| nhlh2 | 0.218 | lbr | -0.107 |
| pbx1a | 0.214 | rpa3 | -0.106 |
| slc32a1 | 0.211 | dut | -0.104 |
| AL845362.1 | 0.204 | tuba8l | -0.103 |
| tfap2c | 0.199 | banf1 | -0.103 |
| nfasca | 0.195 | tuba8l4 | -0.102 |
| pax10 | 0.194 | nutf2l | -0.102 |
| rbfox1 | 0.188 | ccnd1 | -0.101 |
| pbx1b | 0.185 | syt5b | -0.101 |
| calb2a | 0.181 | cks1b | -0.101 |
| tfap2a | 0.179 | crx | -0.100 |
| stxbp1a | 0.178 | fen1 | -0.100 |
| hmgb3a | 0.178 | lig1 | -0.099 |
| rbfox2 | 0.177 | id1 | -0.098 |
| syt2a | 0.176 | mcm7 | -0.096 |
| grin1a | 0.176 | anp32e | -0.096 |
| stx1b | 0.174 | si:ch73-281n10.2 | -0.095 |
| gabrb2 | 0.163 | otx5 | -0.095 |
| eno2 | 0.162 | mibp | -0.094 |
| zgc:92912 | 0.161 | mad2l1 | -0.094 |
| tspan18b | 0.160 | dlgap5 | -0.093 |
| ttyh2 | 0.159 | rpa2 | -0.092 |
| rtn1a | 0.158 | nasp | -0.092 |
| gpm6aa | 0.158 | aurkb | -0.091 |
| tubb5 | 0.156 | mcm6 | -0.091 |
| tfap2b | 0.153 | cldn5a | -0.090 |