"sorting and assembly machinery component 50 homolog, like"
ZFIN 
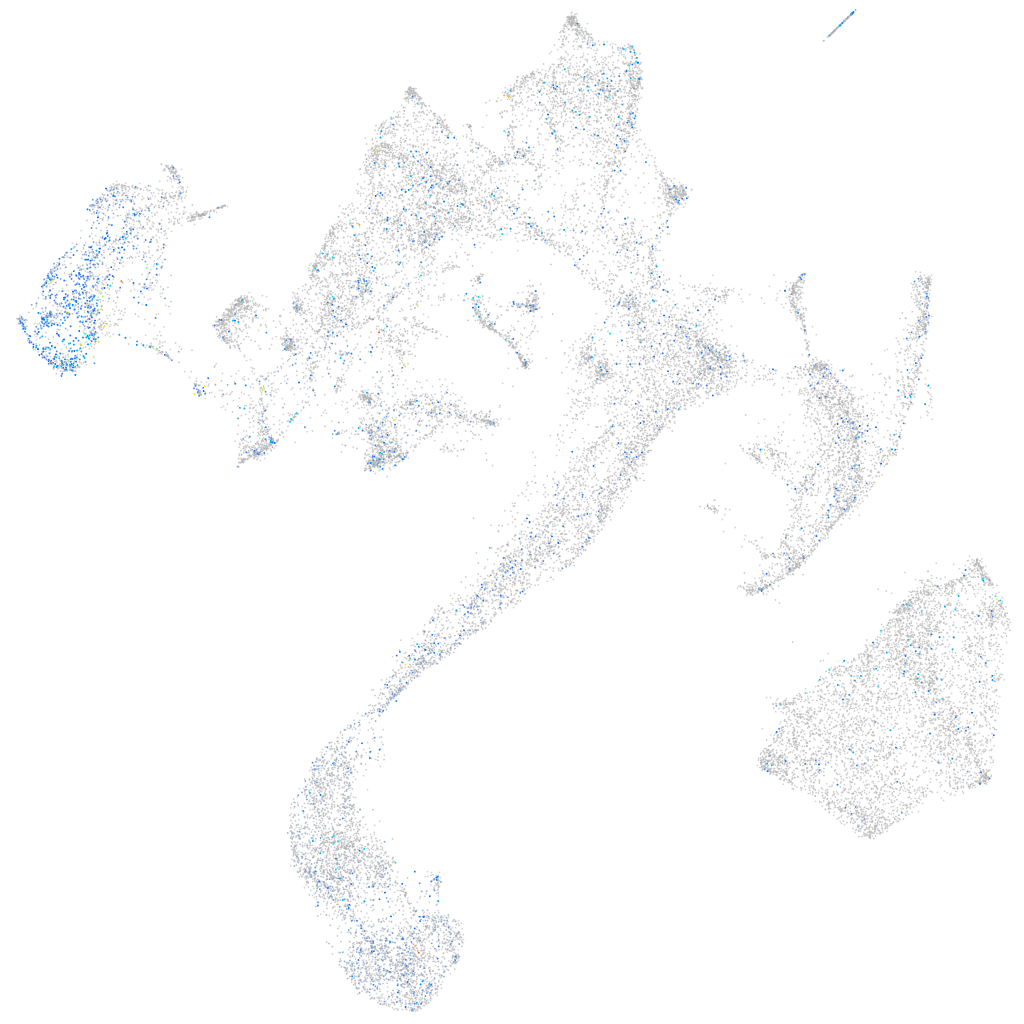
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mybpc3 | 0.268 | hsp90ab1 | -0.171 |
| tnnt2d | 0.260 | eef1a1l1 | -0.165 |
| tnni4b.2 | 0.259 | h3f3d | -0.164 |
| tnnt2e | 0.256 | pabpc1a | -0.154 |
| hhatlb | 0.253 | cirbpb | -0.153 |
| LO018250.1 | 0.246 | h2afvb | -0.153 |
| pvalb4 | 0.245 | ran | -0.152 |
| tnnc1b | 0.245 | khdrbs1a | -0.150 |
| BX248497.1 | 0.236 | hnrnpaba | -0.148 |
| CU633479.2 | 0.230 | si:ch73-1a9.3 | -0.148 |
| ryr1a | 0.229 | naca | -0.147 |
| myoz2b | 0.227 | hmgb2b | -0.144 |
| got1 | 0.226 | hmga1a | -0.142 |
| aco2 | 0.225 | hnrnpabb | -0.141 |
| myom1b | 0.225 | si:ch73-281n10.2 | -0.139 |
| smyhc1 | 0.223 | cirbpa | -0.139 |
| ugp2a | 0.221 | hnrnpa0b | -0.138 |
| ckmt2b | 0.220 | si:ch211-222l21.1 | -0.136 |
| glud1b | 0.216 | npm1a | -0.136 |
| zgc:154046 | 0.213 | setb | -0.135 |
| cs | 0.212 | syncrip | -0.133 |
| prx | 0.212 | snrpf | -0.133 |
| nnt | 0.211 | cbx3a | -0.133 |
| pygmb | 0.211 | snrpb | -0.133 |
| myl13 | 0.209 | hmgb2a | -0.131 |
| tnni1c | 0.209 | ncl | -0.130 |
| nme2b.2 | 0.208 | nop58 | -0.129 |
| mdh2 | 0.208 | snrpd1 | -0.128 |
| eno3 | 0.208 | snrpd2 | -0.127 |
| tpm2 | 0.208 | ddx39ab | -0.127 |
| fxyd1 | 0.206 | ptmab | -0.126 |
| atp5if1b | 0.206 | anp32b | -0.125 |
| casq2 | 0.204 | nop56 | -0.125 |
| gapdh | 0.203 | snu13b | -0.125 |
| rtn1a | 0.201 | hmgn7 | -0.124 |