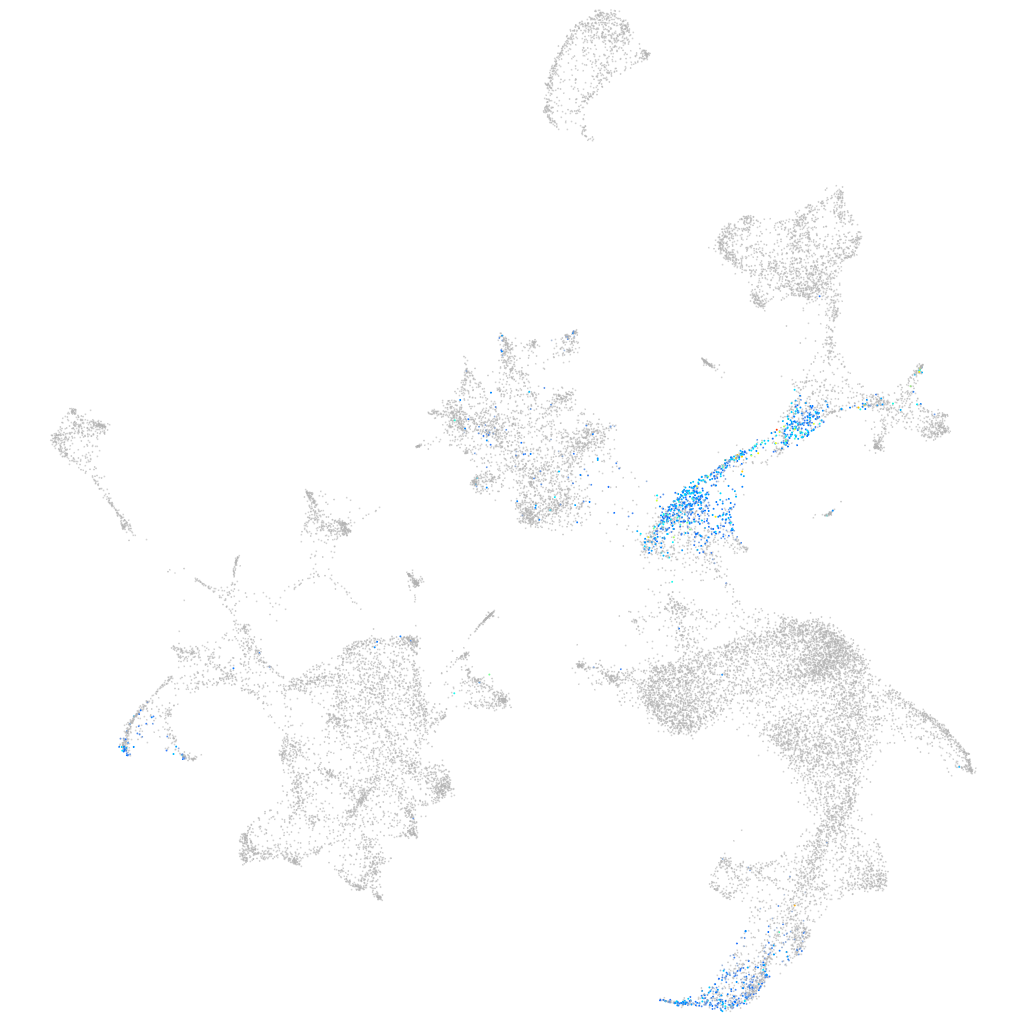
sphingosine-1-phosphate receptor 5b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 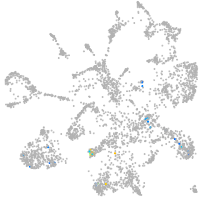 |
mesenchyme 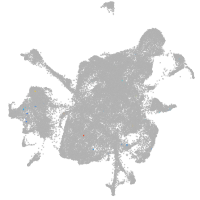 |
hematopoietic / vasculature 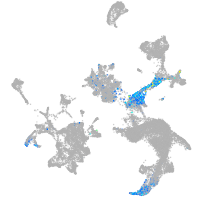 |
pronephros 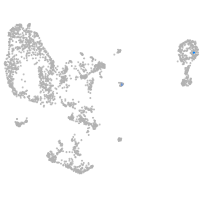 |
neural |
spinal cord / glia |
eye |
taste / olfactory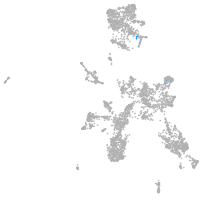 |
otic / lateral line |
epidermis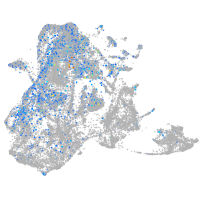 |
periderm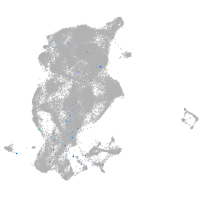 |
fin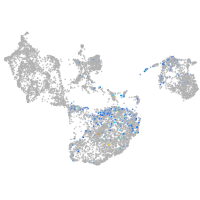 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myb | 0.443 | ctsla | -0.129 |
| blf | 0.385 | marcksl1b | -0.124 |
| hdac7b | 0.314 | gabarapa | -0.109 |
| ahcy | 0.305 | fthl27 | -0.105 |
| si:dkey-261j4.4 | 0.301 | krt8 | -0.102 |
| hdr | 0.292 | marcksl1a | -0.101 |
| suv39h1b | 0.291 | krt18a.1 | -0.100 |
| AK6 | 0.289 | bloc1s6 | -0.099 |
| dachb | 0.285 | id1 | -0.099 |
| snu13b | 0.282 | si:ch211-250g4.3 | -0.097 |
| c1qbp | 0.280 | cd81a | -0.096 |
| fermt3b | 0.280 | hbae3 | -0.095 |
| nop58 | 0.275 | ccni | -0.094 |
| aamp | 0.272 | akap12b | -0.092 |
| pa2g4a | 0.271 | tpm4a | -0.091 |
| npm1a | 0.269 | zgc:92606 | -0.091 |
| spi2 | 0.260 | cdh5 | -0.090 |
| gtpbp4 | 0.260 | hbbe1.3 | -0.087 |
| nop56 | 0.259 | cnn2 | -0.086 |
| bysl | 0.256 | si:ch211-156j16.1 | -0.084 |
| eif6 | 0.256 | cpn1 | -0.084 |
| riox2 | 0.256 | cahz | -0.084 |
| polr1d | 0.255 | vat1 | -0.083 |
| nop10 | 0.254 | etv2 | -0.083 |
| cd3eap | 0.254 | si:dkey-56m19.5 | -0.082 |
| myg1 | 0.251 | hbbe1.1 | -0.082 |
| si:ch211-214p16.2 | 0.249 | gabarapb | -0.082 |
| tsr2 | 0.249 | egfl7 | -0.082 |
| si:dkey-102m7.3 | 0.249 | clec14a | -0.082 |
| drl | 0.249 | rhoca | -0.081 |
| ddx21 | 0.248 | cldn5b | -0.080 |
| men1 | 0.245 | she | -0.080 |
| eif4ebp3l | 0.245 | nova2 | -0.080 |
| mrpl12 | 0.244 | tagln2 | -0.079 |
| tcp1 | 0.244 | anxa3b | -0.079 |