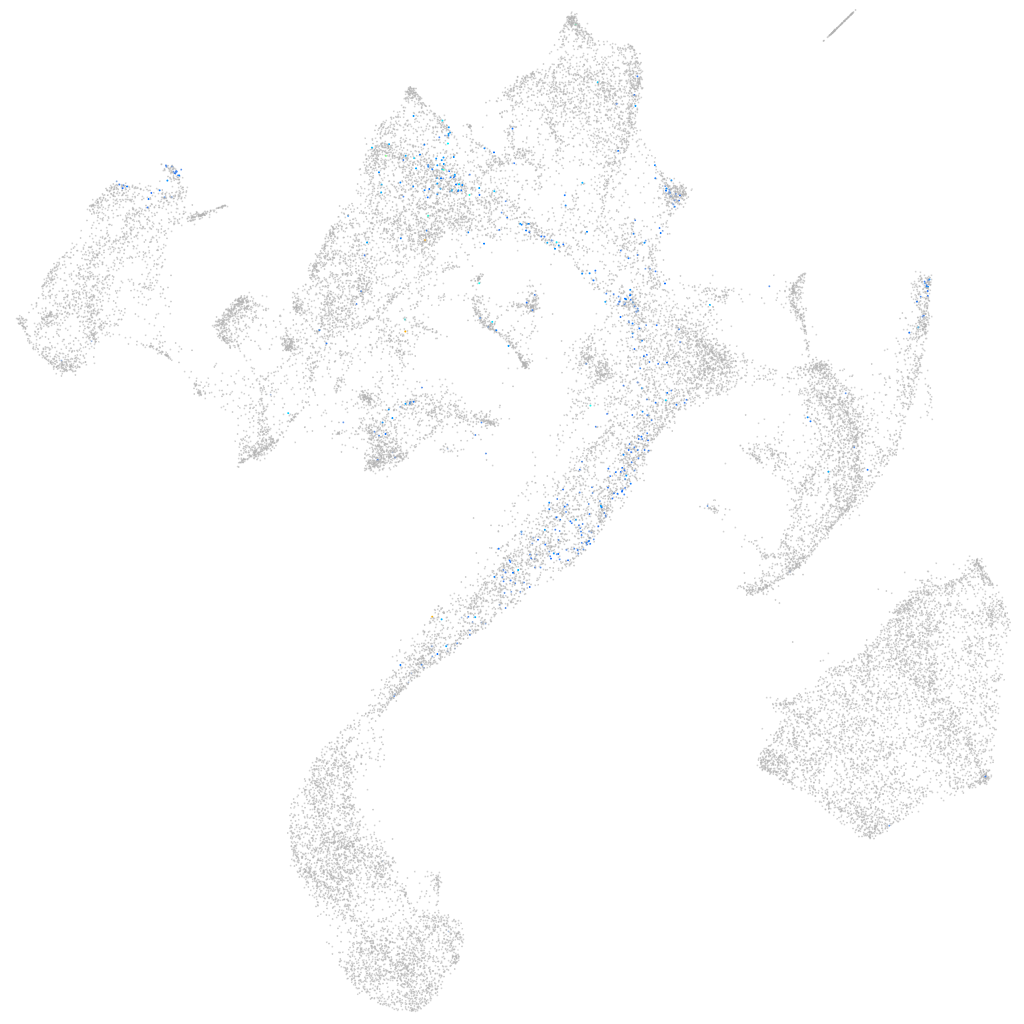
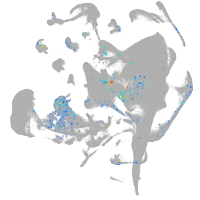
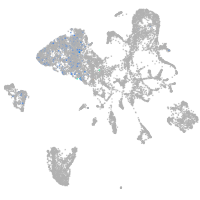
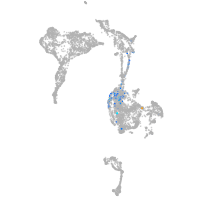
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX000438.1 | 0.130 | cox7c | -0.057 |
| si:ch211-43f4.1 | 0.125 | cdx4 | -0.055 |
| LOC108183588 | 0.118 | hes6 | -0.053 |
| emp2 | 0.115 | apoc1 | -0.048 |
| rbp7b | 0.111 | tnnt3b | -0.048 |
| CR354547.3 | 0.108 | pvalb1 | -0.047 |
| v2rh7 | 0.106 | wnt5b | -0.047 |
| slc1a3a | 0.105 | prx | -0.046 |
| vwde | 0.102 | eno3 | -0.046 |
| fstl1a | 0.101 | si:ch73-367p23.2 | -0.046 |
| glula | 0.100 | pvalb2 | -0.046 |
| cd81a | 0.097 | si:ch211-255p10.3 | -0.045 |
| jam2a | 0.091 | tbx16 | -0.045 |
| lman1 | 0.090 | casq1b | -0.045 |
| lgi2a | 0.090 | sp5l | -0.045 |
| peli1b | 0.088 | pkmb | -0.044 |
| trabd2a | 0.088 | tnni2a.4 | -0.044 |
| CABZ01118738.1 | 0.087 | apoeb | -0.043 |
| smoc1 | 0.086 | si:ch211-152c2.3 | -0.043 |
| fbln7 | 0.086 | vox | -0.043 |
| CABZ01075068.1 | 0.084 | pgam2 | -0.043 |
| si:ch1073-268j14.1 | 0.083 | atp5md | -0.043 |
| mdka | 0.083 | atp5f1e | -0.043 |
| ssr3 | 0.083 | cdx1a | -0.043 |
| thbs4b | 0.083 | rtn1a | -0.042 |
| sparc | 0.083 | gapdh | -0.042 |
| col6a2 | 0.082 | cavin4a | -0.042 |
| col6a3 | 0.080 | si:dkey-16p21.8 | -0.042 |
| thrap3a | 0.079 | dhrs7cb | -0.042 |
| midn | 0.079 | ckmb | -0.041 |
| cygb1 | 0.079 | msgn1 | -0.041 |
| cilp | 0.079 | tnnt3a | -0.041 |
| tcf12 | 0.078 | CABZ01061524.1 | -0.041 |
| rgmd | 0.078 | myom1a | -0.041 |
| col5a1 | 0.078 | slc25a4 | -0.040 |