ribosome binding protein 1b
ZFIN 
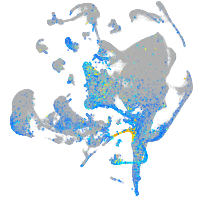
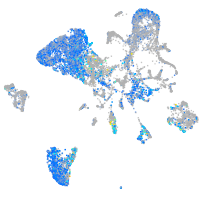
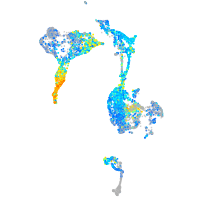
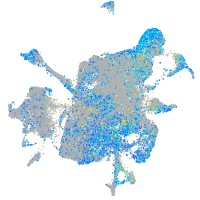
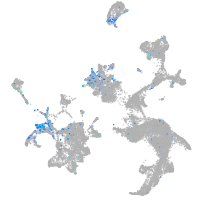
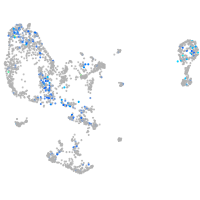
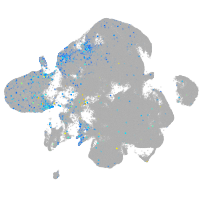
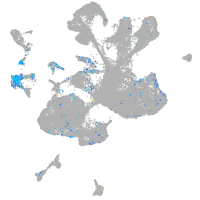
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lft2 | 0.183 | tuba1c | -0.106 |
| lft1 | 0.120 | ptmaa | -0.105 |
| shhb | 0.109 | elavl3 | -0.097 |
| cnn2 | 0.109 | rtn1a | -0.093 |
| tagln2 | 0.104 | gpm6aa | -0.088 |
| krt8 | 0.103 | stmn1b | -0.088 |
| fermt1 | 0.102 | nova2 | -0.088 |
| asph | 0.101 | tmsb | -0.078 |
| tpm4a | 0.100 | hmgb3a | -0.078 |
| krt18a.1 | 0.096 | ckbb | -0.075 |
| pmp22a | 0.096 | h3f3a | -0.071 |
| lamb1a | 0.096 | tubb5 | -0.070 |
| arhgef5 | 0.093 | gpm6ab | -0.068 |
| akap12b | 0.092 | gng3 | -0.068 |
| pak2a | 0.091 | si:dkey-276j7.1 | -0.067 |
| cyp2aa6 | 0.090 | tuba1a | -0.067 |
| nkx1.2la | 0.090 | hnrnpa0l | -0.066 |
| serpinh1b | 0.090 | sncb | -0.066 |
| crtap | 0.089 | fabp3 | -0.066 |
| myh9a | 0.089 | rnasekb | -0.064 |
| nid2a | 0.088 | jpt1b | -0.064 |
| hspb1 | 0.087 | marcksl1a | -0.063 |
| fkbp9 | 0.086 | rtn1b | -0.063 |
| nid1b | 0.085 | zc4h2 | -0.062 |
| fn1a | 0.084 | atp6v0cb | -0.062 |
| npm1a | 0.083 | ywhah | -0.061 |
| NC-002333.4 | 0.083 | hmgb1a | -0.061 |
| apoc1 | 0.082 | tubb2b | -0.060 |
| rspo3 | 0.081 | myt1b | -0.060 |
| apoeb | 0.081 | elavl4 | -0.060 |
| pdlim1 | 0.081 | si:ch1073-429i10.3.1 | -0.060 |
| bambia | 0.080 | gng2 | -0.060 |
| zfp36l1a | 0.079 | fam168a | -0.059 |
| sdc4 | 0.079 | myt1a | -0.059 |
| cdh1 | 0.079 | calm1a | -0.059 |