RNA pseudouridine synthase domain containing 2
ZFIN 
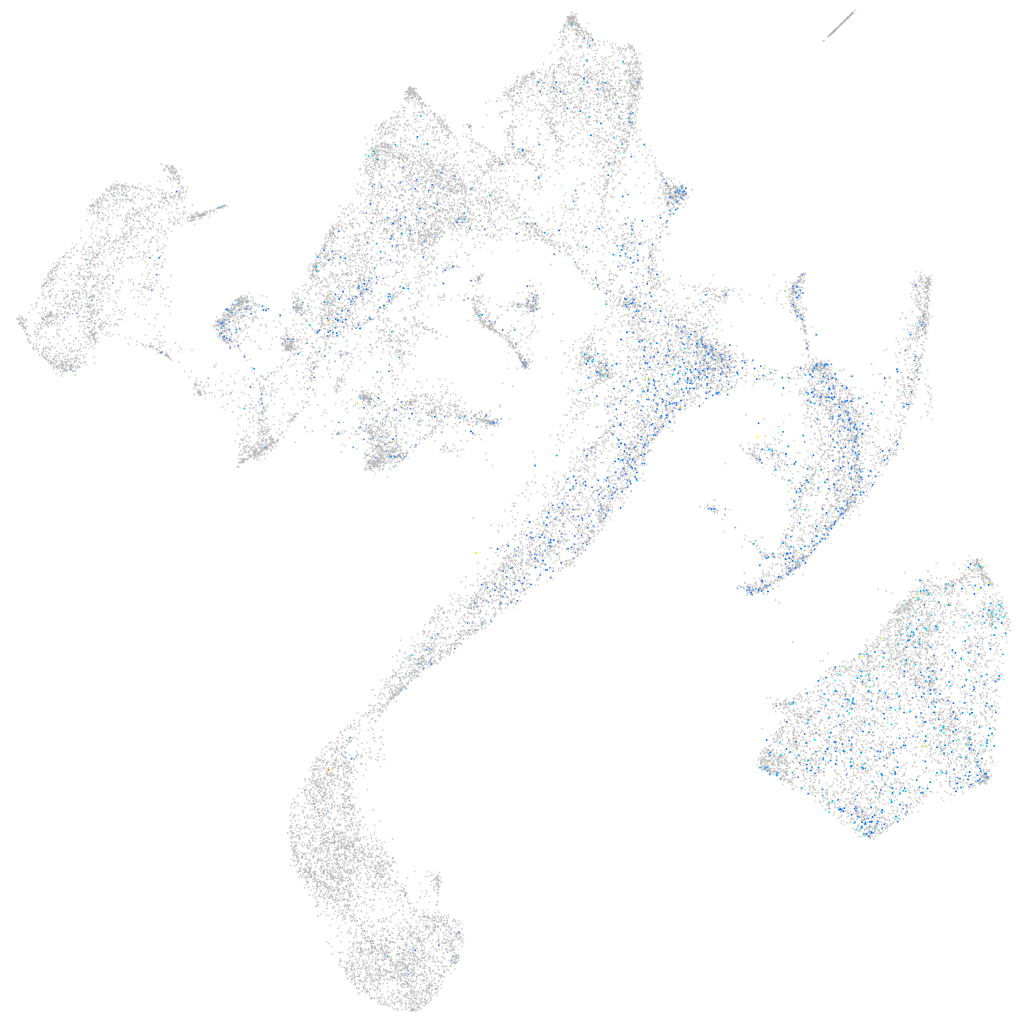
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nop58 | 0.168 | actc1b | -0.115 |
| npm1a | 0.166 | ak1 | -0.111 |
| dkc1 | 0.163 | ckma | -0.109 |
| ncl | 0.160 | ckmb | -0.108 |
| nop56 | 0.158 | neb | -0.107 |
| hnrnpabb | 0.158 | tnnc2 | -0.106 |
| setb | 0.156 | eno3 | -0.105 |
| snu13b | 0.154 | atp2a1 | -0.104 |
| fbl | 0.152 | ldb3b | -0.103 |
| nop2 | 0.149 | actn3a | -0.103 |
| nhp2 | 0.147 | aldoab | -0.103 |
| prmt1 | 0.146 | si:ch73-367p23.2 | -0.102 |
| cbx3a | 0.144 | smyd1a | -0.101 |
| hnrnpaba | 0.144 | acta1b | -0.101 |
| khdrbs1a | 0.143 | bhmt | -0.100 |
| ptges3b | 0.143 | nme2b.2 | -0.099 |
| syncrip | 0.142 | pvalb1 | -0.099 |
| anp32b | 0.141 | gapdh | -0.099 |
| rsl1d1 | 0.141 | mylpfa | -0.098 |
| hmga1a | 0.141 | tpma | -0.098 |
| pprc1 | 0.140 | tpi1b | -0.097 |
| nop10 | 0.140 | si:ch211-266g18.10 | -0.097 |
| snrpd1 | 0.140 | eno1a | -0.097 |
| twistnb | 0.139 | actn3b | -0.097 |
| cirbpa | 0.139 | si:dkey-16p21.8 | -0.097 |
| rbm8a | 0.138 | tnnt3a | -0.097 |
| ddx39ab | 0.138 | pvalb2 | -0.097 |
| lyar | 0.138 | myom1a | -0.096 |
| srsf1a | 0.138 | casq2 | -0.096 |
| gar1 | 0.137 | pmp22b | -0.096 |
| snrpb | 0.137 | vdac3 | -0.095 |
| snrpe | 0.137 | XLOC-025819 | -0.095 |
| hnrnpa0b | 0.137 | tmod4 | -0.095 |
| wdr43 | 0.136 | ldb3a | -0.094 |
| ebna1bp2 | 0.135 | pgam2 | -0.094 |