ribosomal protein L27a
ZFIN 
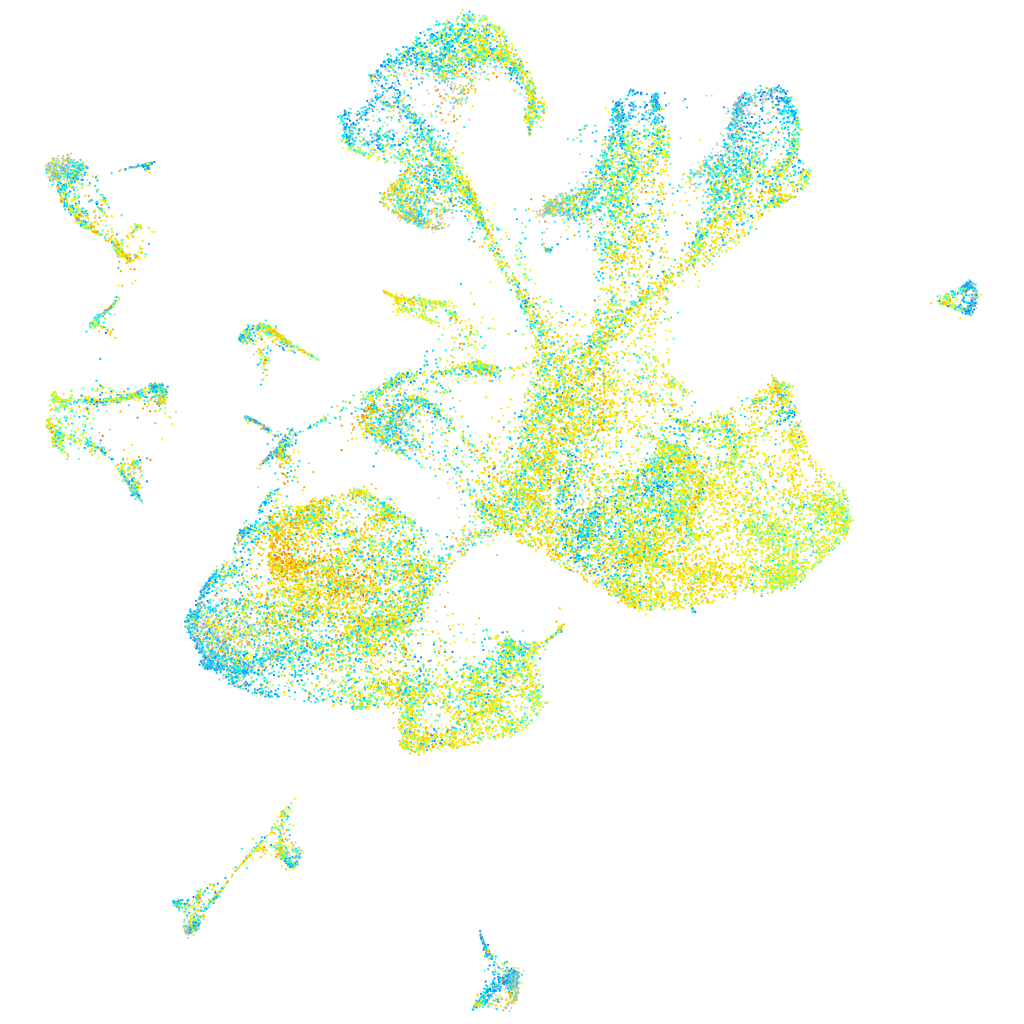
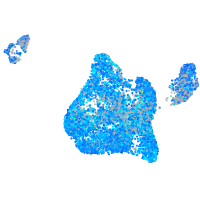
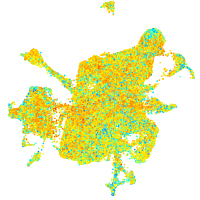
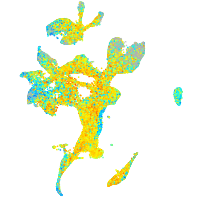
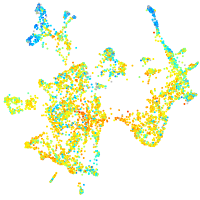
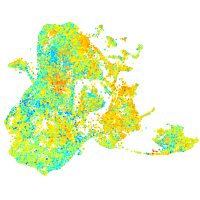
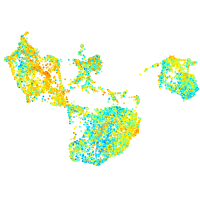
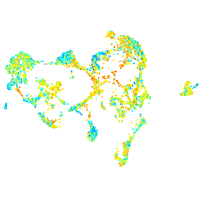
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rps7 | 0.484 | acbd7 | -0.276 |
| rps20 | 0.479 | aldocb | -0.275 |
| rps5 | 0.476 | gapdhs | -0.267 |
| rplp2l | 0.476 | glula | -0.264 |
| rps12 | 0.473 | CR383676.1 | -0.258 |
| rplp1 | 0.472 | cx43 | -0.244 |
| rpl23 | 0.472 | sncgb | -0.237 |
| rpl13 | 0.468 | elavl4 | -0.234 |
| rpl26 | 0.467 | mt2 | -0.234 |
| rps15a | 0.467 | ndrg3a | -0.233 |
| rpl7a | 0.467 | snap25a | -0.231 |
| rps13 | 0.466 | efhd1 | -0.228 |
| rps14 | 0.464 | IGLON5 | -0.226 |
| rpl12 | 0.464 | slc4a4a | -0.223 |
| rpl21 | 0.464 | atp6v0cb | -0.216 |
| rpl8 | 0.461 | calm1a | -0.215 |
| rpl10a | 0.460 | ckbb | -0.214 |
| rps24 | 0.459 | slc6a1b | -0.213 |
| rps23 | 0.458 | eno2 | -0.213 |
| rpl11 | 0.458 | vsnl1b | -0.213 |
| rpl17 | 0.455 | pygmb | -0.212 |
| rpl9 | 0.455 | atp2b3a | -0.212 |
| rps9 | 0.455 | eno1a | -0.212 |
| si:dkey-151g10.6 | 0.455 | tpi1b | -0.207 |
| rps10 | 0.451 | gpm6ab | -0.206 |
| rps19 | 0.448 | map4l | -0.205 |
| rps3a | 0.448 | calm3a | -0.205 |
| rpl19 | 0.447 | ak1 | -0.203 |
| rpsa | 0.446 | stx1b | -0.203 |
| rpl27 | 0.446 | spock2 | -0.202 |
| rps27.1 | 0.446 | calm1b | -0.201 |
| rps27a | 0.445 | pvalb2 | -0.201 |
| rps2 | 0.444 | si:ch73-119p20.1 | -0.200 |
| rpl7 | 0.444 | pvalb1 | -0.200 |
| rpl10 | 0.443 | cadm4 | -0.200 |