receptor tyrosine kinase-like orphan receptor 1
ZFIN 
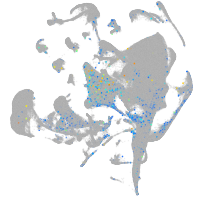
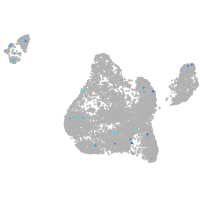
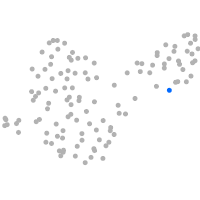
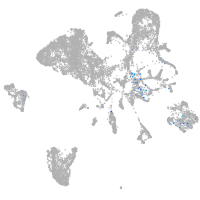
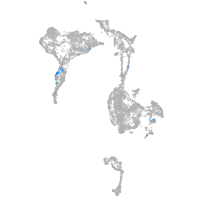
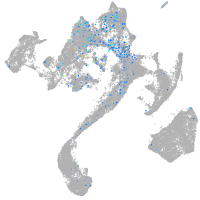
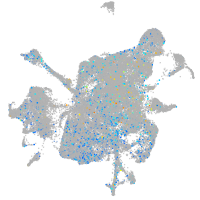
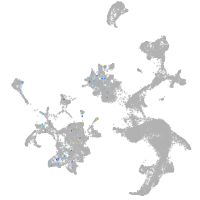
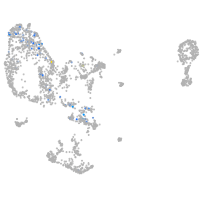
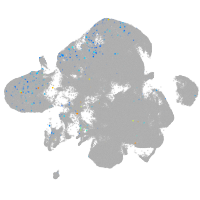
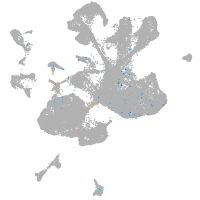
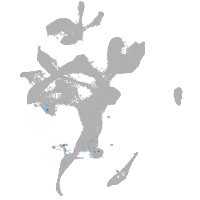
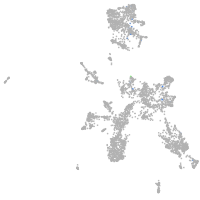
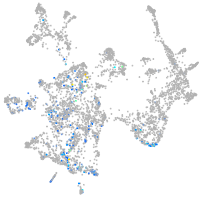
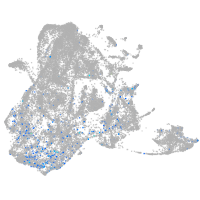
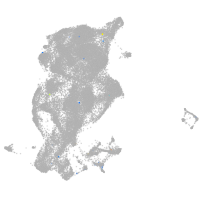
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX927258.1 | 0.058 | tuba1c | -0.056 |
| dkc1 | 0.056 | ptmaa | -0.054 |
| npm1a | 0.056 | gpm6aa | -0.052 |
| nop58 | 0.055 | rtn1a | -0.051 |
| hspb1 | 0.055 | elavl3 | -0.051 |
| rsl1d1 | 0.054 | stmn1b | -0.049 |
| fbl | 0.053 | ckbb | -0.044 |
| lig1 | 0.052 | tmsb | -0.041 |
| XLOC-032526 | 0.052 | hmgb3a | -0.038 |
| nop56 | 0.051 | gng3 | -0.038 |
| zic2b | 0.051 | tubb5 | -0.038 |
| ebna1bp2 | 0.050 | nova2 | -0.037 |
| nop2 | 0.050 | gpm6ab | -0.037 |
| id1 | 0.050 | rnasekb | -0.037 |
| si:ch211-152c2.3 | 0.050 | tmsb4x | -0.037 |
| zfp36l1a | 0.050 | sncb | -0.036 |
| nr6a1a | 0.049 | marcksl1a | -0.034 |
| snu13b | 0.049 | atp6v0cb | -0.034 |
| akap12b | 0.049 | ywhah | -0.034 |
| gnl3 | 0.049 | fez1 | -0.034 |
| nhp2 | 0.049 | myt1b | -0.034 |
| ncl | 0.049 | gapdhs | -0.034 |
| cad | 0.048 | elavl4 | -0.034 |
| NC-002333.4 | 0.048 | h3f3c | -0.034 |
| rrp1 | 0.048 | atp6v1g1 | -0.034 |
| aldob | 0.048 | zc4h2 | -0.033 |
| BX248410.1 | 0.048 | rtn1b | -0.033 |
| bms1 | 0.047 | ppdpfb | -0.033 |
| si:dkey-102m7.3 | 0.047 | celf2 | -0.033 |
| si:dkey-239h2.3 | 0.047 | vamp2 | -0.033 |
| pprc1 | 0.047 | mllt11 | -0.033 |
| apoeb | 0.046 | atp6v1e1b | -0.032 |
| eif5a2 | 0.046 | h3f3a | -0.032 |
| mybbp1a | 0.046 | si:ch1073-429i10.3.1 | -0.032 |
| pes | 0.046 | calm1a | -0.032 |