"ribonuclease, RNase K b"
ZFIN 
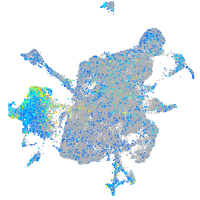
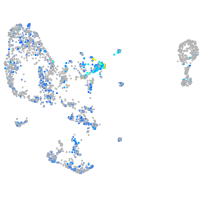
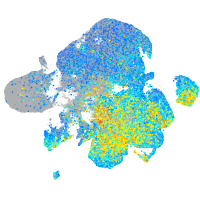
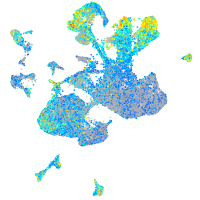
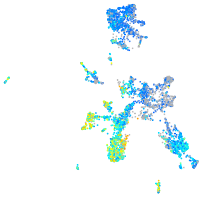
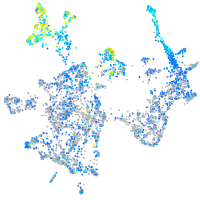
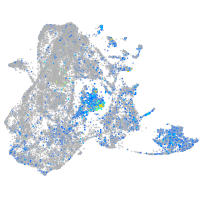
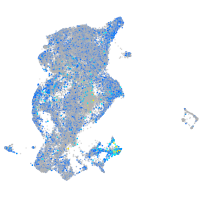
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.479 | gapdh | -0.213 |
| neurod1 | 0.434 | gamt | -0.213 |
| pcsk1nl | 0.421 | gatm | -0.198 |
| gapdhs | 0.421 | ahcy | -0.190 |
| syt1a | 0.412 | fbp1b | -0.181 |
| vamp2 | 0.404 | mat1a | -0.169 |
| scgn | 0.389 | gstr | -0.168 |
| calm1b | 0.389 | aldob | -0.163 |
| mir7a-1 | 0.387 | gstt1a | -0.162 |
| atp6v0cb | 0.380 | cx32.3 | -0.162 |
| pax6b | 0.375 | nupr1b | -0.161 |
| tspan7b | 0.372 | aqp12 | -0.161 |
| si:dkey-153k10.9 | 0.369 | scp2a | -0.158 |
| insm1a | 0.365 | apoa4b.1 | -0.157 |
| id4 | 0.361 | bhmt | -0.157 |
| vat1 | 0.360 | pnp4b | -0.153 |
| mir375-2 | 0.358 | apoc2 | -0.153 |
| pcsk1 | 0.356 | aldh6a1 | -0.150 |
| aldocb | 0.350 | apoc1 | -0.150 |
| gnao1a | 0.349 | glud1b | -0.150 |
| scg2a | 0.346 | aldh7a1 | -0.147 |
| necap1 | 0.342 | gcshb | -0.146 |
| isl1 | 0.341 | afp4 | -0.146 |
| atpv0e2 | 0.340 | sult2st2 | -0.145 |
| rprmb | 0.334 | cox7a1 | -0.145 |
| egr4 | 0.334 | rdh1 | -0.144 |
| ptmaa | 0.333 | hdlbpa | -0.144 |
| cpe | 0.332 | agxtb | -0.144 |
| myt1b | 0.331 | hspe1 | -0.141 |
| fev | 0.330 | slc16a10 | -0.141 |
| zgc:101731 | 0.329 | abat | -0.141 |
| snap25a | 0.328 | apoa1b | -0.140 |
| calm1a | 0.327 | npm1a | -0.136 |
| kcnj11 | 0.327 | agxta | -0.136 |
| gpc1a | 0.326 | si:dkey-16p21.8 | -0.135 |