"ribonuclease, RNase K a"
ZFIN 
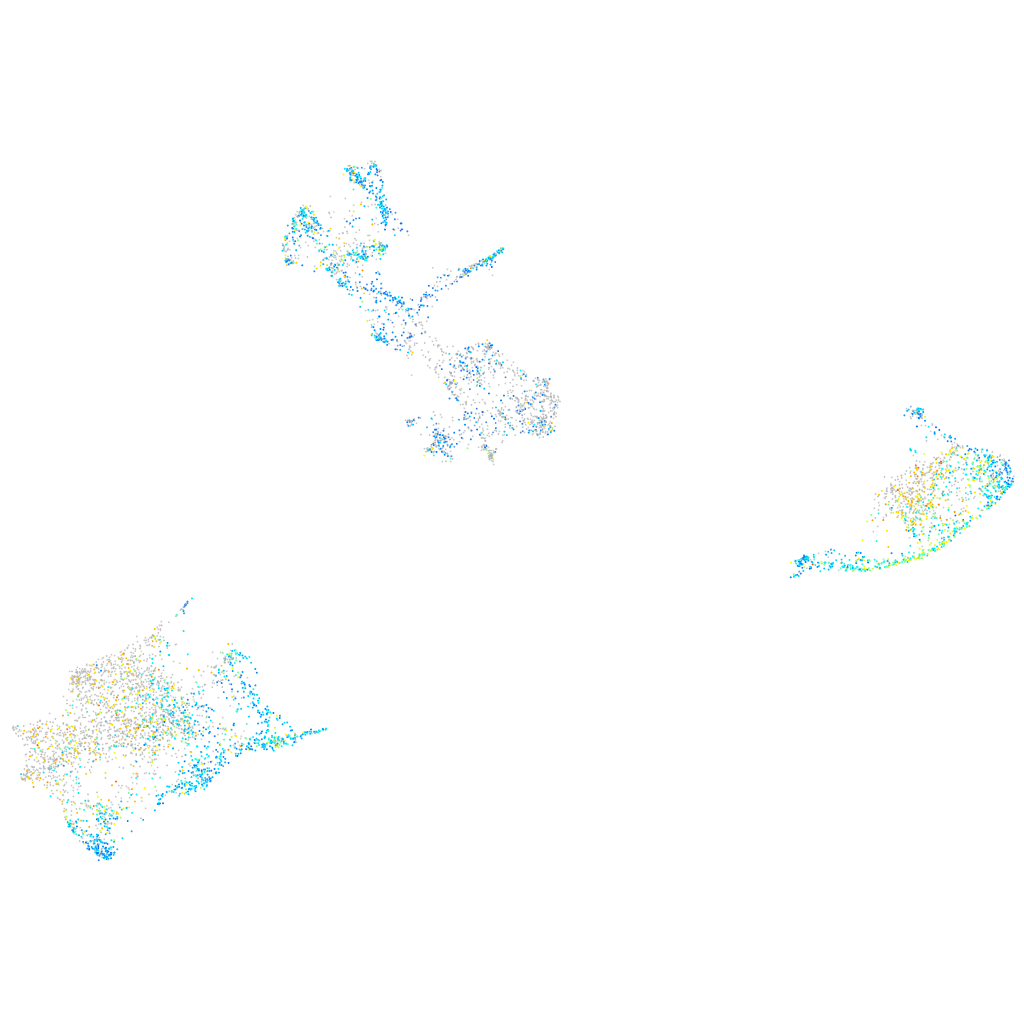
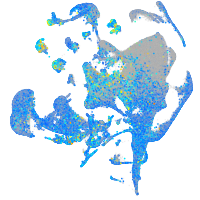
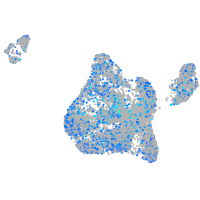
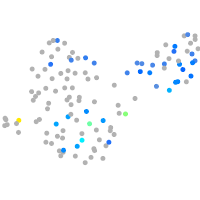
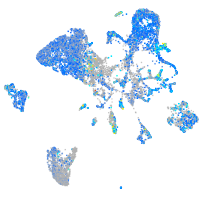
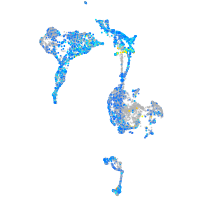
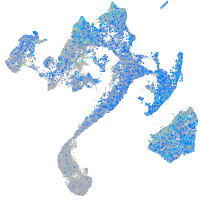
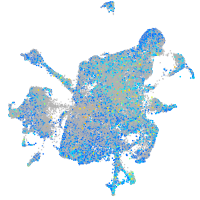
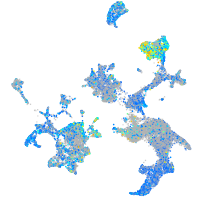
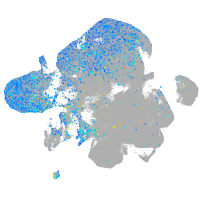
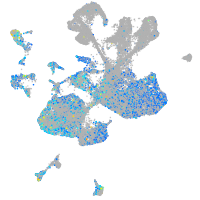
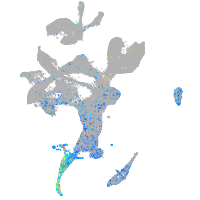
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.337 | ptmaa | -0.208 |
| pttg1ipb | 0.327 | si:ch211-222l21.1 | -0.193 |
| syngr1a | 0.325 | tuba1c | -0.189 |
| degs1 | 0.306 | gpm6aa | -0.181 |
| slc45a2 | 0.284 | nova2 | -0.176 |
| rab32a | 0.283 | elavl3 | -0.173 |
| gpr143 | 0.283 | tmsb | -0.167 |
| sypl2b | 0.283 | stmn1b | -0.161 |
| ctsla | 0.282 | gng3 | -0.156 |
| bace2 | 0.281 | si:ch73-1a9.3 | -0.153 |
| slc3a2a | 0.277 | epb41a | -0.151 |
| slc37a2 | 0.276 | hmgb1b | -0.148 |
| zgc:110239 | 0.274 | sncb | -0.145 |
| atp6ap1b | 0.274 | hmgb3a | -0.143 |
| rab38 | 0.270 | CU467822.1 | -0.142 |
| sdc4 | 0.269 | fez1 | -0.139 |
| scpep1 | 0.267 | ptmab | -0.138 |
| tmem243b | 0.265 | zc4h2 | -0.133 |
| naga | 0.262 | hbae3 | -0.131 |
| tfap2e | 0.261 | sox11b | -0.130 |
| atp6ap2 | 0.260 | cadm3 | -0.129 |
| ctsba | 0.259 | hbbe1.3 | -0.124 |
| rabl6b | 0.257 | elavl4 | -0.124 |
| agtrap | 0.254 | gng2 | -0.123 |
| xbp1 | 0.253 | tmeff1b | -0.123 |
| si:zfos-943e10.1 | 0.253 | pvalb1 | -0.120 |
| tspan10 | 0.252 | stmn1a | -0.118 |
| mitfa | 0.251 | marcksl1a | -0.118 |
| abracl | 0.251 | rbfox1 | -0.118 |
| zgc:165573 | 0.247 | fabp7a | -0.116 |
| dct | 0.246 | FO082781.1 | -0.116 |
| pcdh10a | 0.245 | pvalb2 | -0.116 |
| smim29 | 0.245 | vamp2 | -0.115 |
| cst14a.2 | 0.245 | foxp4 | -0.115 |
| trpm1b | 0.244 | mllt11 | -0.114 |