ribonuclease H1
ZFIN 
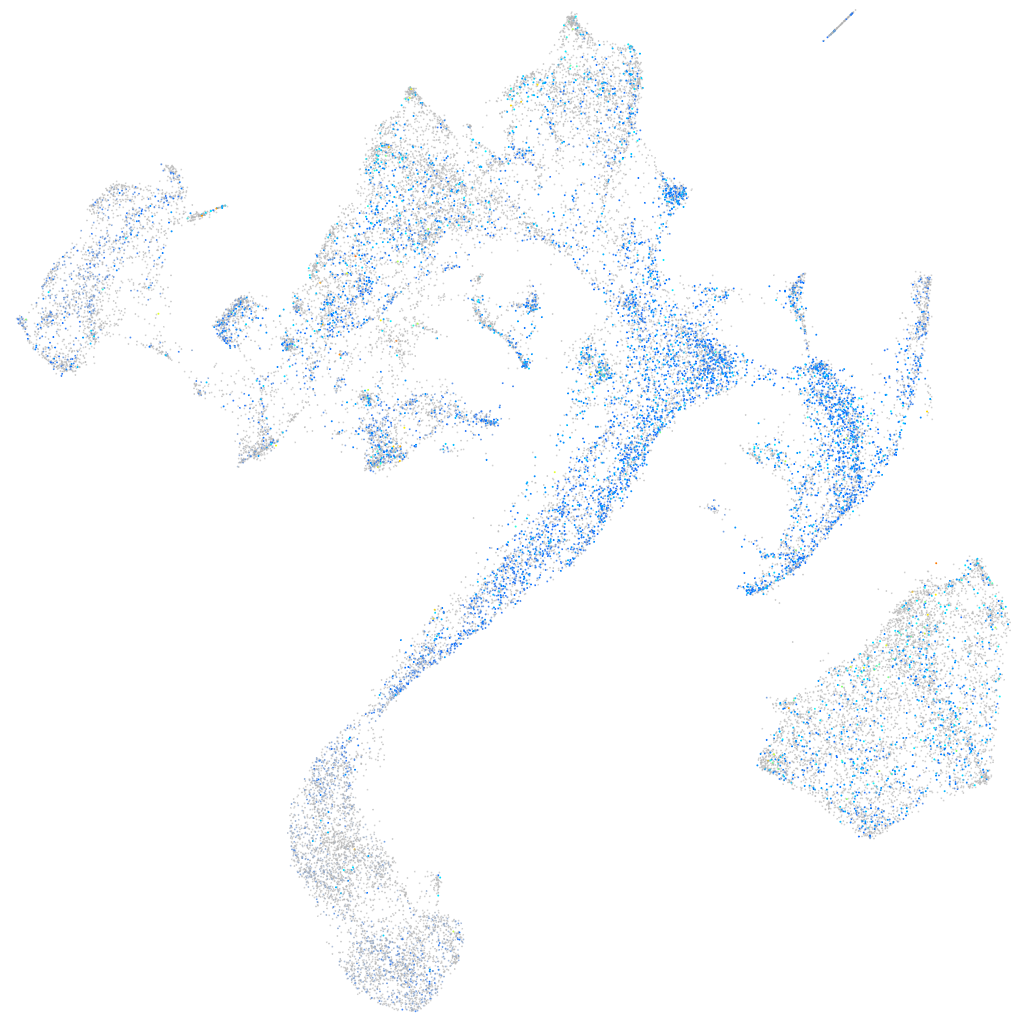
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.212 | pvalb1 | -0.138 |
| tomm20a | 0.204 | ckma | -0.135 |
| cnbpa | 0.202 | actn3a | -0.135 |
| nop10 | 0.200 | si:dkey-16p21.8 | -0.134 |
| rpl7l1 | 0.200 | si:ch73-367p23.2 | -0.134 |
| ran | 0.198 | pvalb2 | -0.133 |
| nhp2 | 0.198 | neb | -0.133 |
| eif4a1a | 0.196 | tnnt3b | -0.131 |
| tma16 | 0.194 | tnnc2 | -0.130 |
| setb | 0.192 | myom1a | -0.130 |
| hnrnpa0l | 0.191 | si:ch211-266g18.10 | -0.130 |
| eif3k | 0.190 | ckmb | -0.129 |
| nifk | 0.188 | smyd1a | -0.128 |
| snrpd2 | 0.188 | XLOC-025819 | -0.128 |
| npm1a | 0.188 | tmod4 | -0.126 |
| cdkn2aipnl | 0.187 | ldb3b | -0.126 |
| bysl | 0.185 | XLOC-001975 | -0.125 |
| snrpf | 0.184 | atp2a1 | -0.125 |
| cct7 | 0.183 | tnni2a.4 | -0.125 |
| eif3ba | 0.182 | ak1 | -0.124 |
| emg1 | 0.182 | XLOC-005350 | -0.124 |
| mrto4 | 0.181 | gapdh | -0.123 |
| CABZ01068251.1 | 0.180 | actn3b | -0.123 |
| cct3 | 0.180 | prx | -0.123 |
| si:dkey-23i12.5 | 0.180 | mylz3 | -0.122 |
| tsr2 | 0.180 | si:ch211-255p10.3 | -0.122 |
| pbdc1 | 0.180 | XLOC-006515 | -0.121 |
| ddx39ab | 0.180 | CABZ01061524.1 | -0.121 |
| ptges3b | 0.179 | tnnt3a | -0.121 |
| eif3s6ip | 0.179 | bhmt | -0.120 |
| naca | 0.179 | mylpfb | -0.120 |
| psma5 | 0.179 | mylpfa | -0.119 |
| btf3l4 | 0.179 | actc1b | -0.118 |
| npm3 | 0.178 | eno3 | -0.118 |
| alyref | 0.178 | pgam2 | -0.117 |