ribosomal modification protein rimK-like family member A
ZFIN 
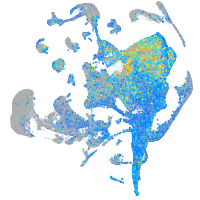
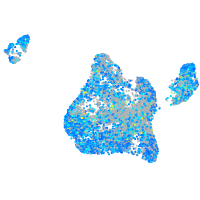
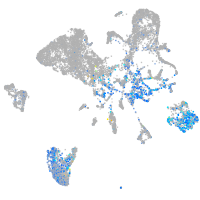
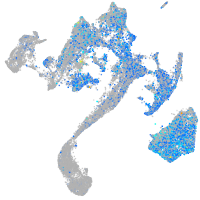
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.215 | si:ch211-139a5.9 | -0.195 |
| si:ch211-137a8.4 | 0.214 | ndufa4l | -0.190 |
| khdrbs1a | 0.213 | cox5ab | -0.186 |
| marcksl1b | 0.208 | cox6a2 | -0.180 |
| hnrnpaba | 0.208 | eno3 | -0.177 |
| hmgb1b | 0.204 | mt-nd1 | -0.175 |
| h3f3d | 0.196 | cdaa | -0.174 |
| si:ch73-281n10.2 | 0.194 | aldh6a1 | -0.171 |
| cbx3a | 0.190 | atp1b1a | -0.170 |
| hmga1a | 0.189 | cdh17 | -0.170 |
| cirbpa | 0.188 | atp5pb | -0.168 |
| cirbpb | 0.186 | rpl37 | -0.167 |
| akap12b | 0.184 | atp5mc1 | -0.167 |
| si:ch73-1a9.3 | 0.184 | cox7a1 | -0.166 |
| CABZ01080702.1 | 0.183 | si:dkey-16p21.8 | -0.164 |
| anp32e | 0.183 | gpx4a | -0.162 |
| cdx4 | 0.182 | mt-nd2 | -0.161 |
| si:dkey-33c12.12 | 0.181 | cox5b2 | -0.160 |
| ptmab | 0.180 | gapdh | -0.160 |
| nat8l | 0.177 | elovl1b | -0.160 |
| cbx5 | 0.176 | atp5l | -0.160 |
| marcksb | 0.175 | atp5mc3b | -0.159 |
| si:ch211-222l21.1 | 0.175 | mt-co1 | -0.159 |
| seta | 0.175 | atp5pf | -0.158 |
| birc5b | 0.174 | COX3 | -0.158 |
| baz1b | 0.173 | sod1 | -0.157 |
| srrm2 | 0.173 | atp5pd | -0.157 |
| hnrnpub | 0.173 | mt-nd5 | -0.156 |
| syncrip | 0.173 | mdh1aa | -0.156 |
| hspb1 | 0.172 | mt-nd4 | -0.155 |
| hnrnpa0b | 0.172 | mt-atp6 | -0.155 |
| hmgn2 | 0.171 | glud1b | -0.154 |
| hmgb2a | 0.171 | ndufa3 | -0.153 |
| tubb2b | 0.170 | slc25a5 | -0.153 |
| neurod4 | 0.169 | eif4a1b | -0.151 |