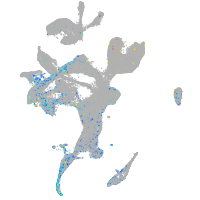
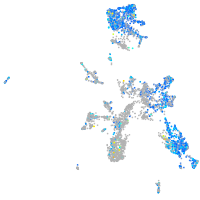
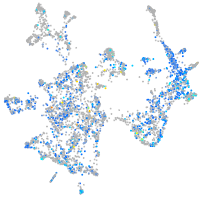
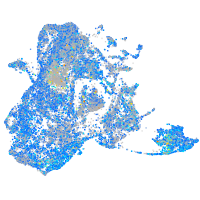
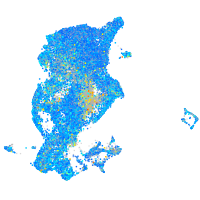
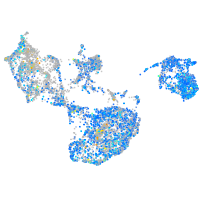
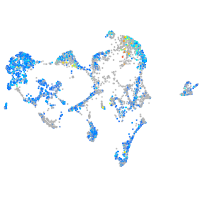
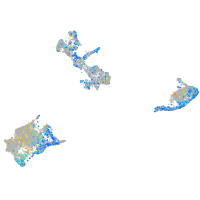
ras homolog family member Ca
ZFIN 
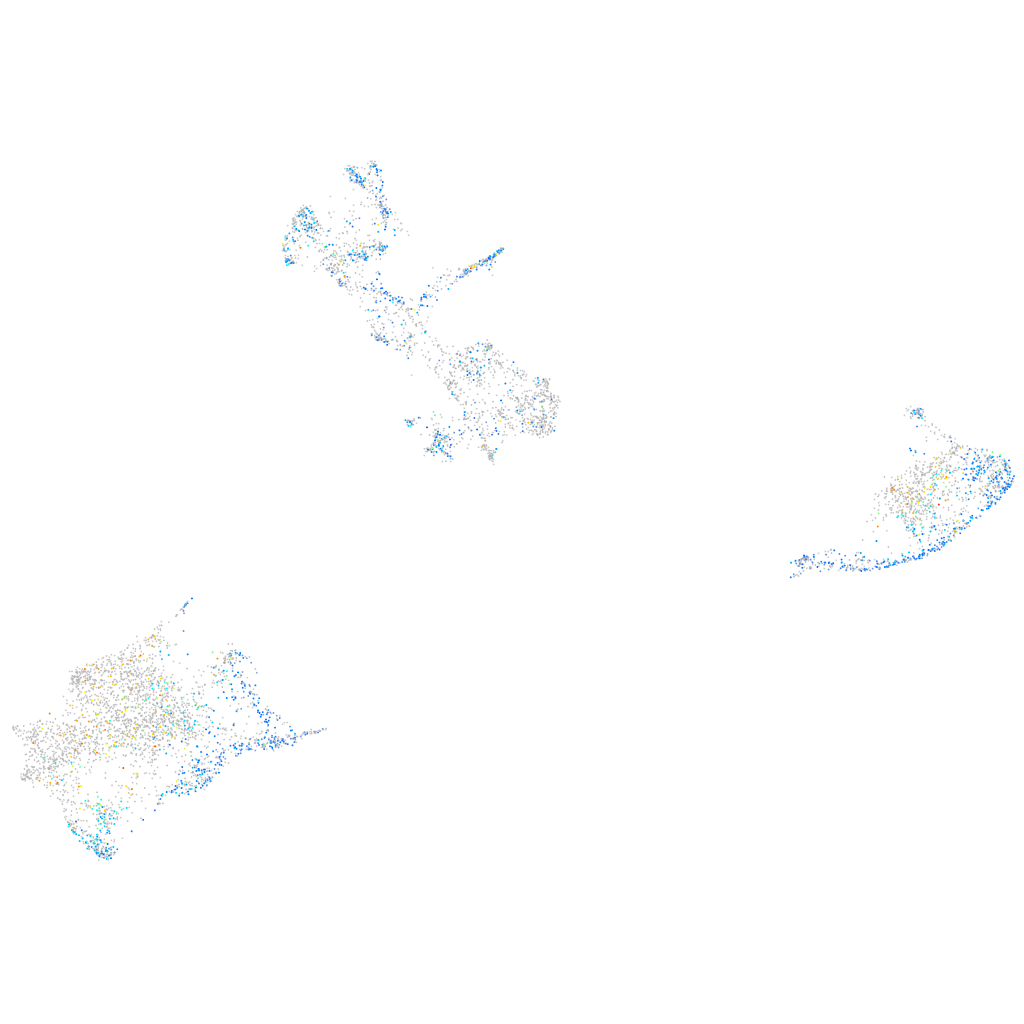
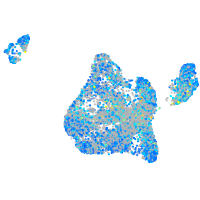
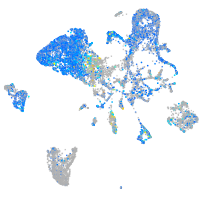
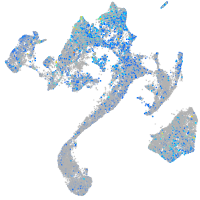
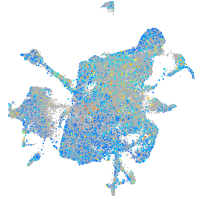
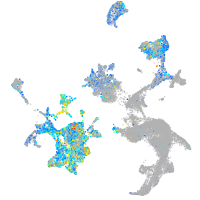
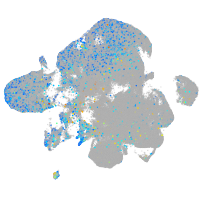
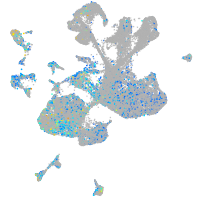
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sdc4 | 0.144 | tuba1c | -0.090 |
| rab32a | 0.126 | si:ch211-222l21.1 | -0.083 |
| tspan36 | 0.126 | stmn1b | -0.081 |
| cst14a.2 | 0.126 | nova2 | -0.080 |
| gpr143 | 0.126 | gpm6aa | -0.079 |
| abracl | 0.124 | elavl3 | -0.076 |
| vamp3 | 0.123 | pvalb1 | -0.073 |
| slc3a2a | 0.123 | gng3 | -0.068 |
| stx12 | 0.121 | tmsb | -0.067 |
| rnaseka | 0.120 | tmeff1b | -0.066 |
| zgc:165573 | 0.119 | ptmaa | -0.065 |
| ap3s1.1 | 0.118 | sox11b | -0.061 |
| syngr1a | 0.115 | CU467822.1 | -0.060 |
| slc45a2 | 0.115 | tox | -0.060 |
| gdi2 | 0.115 | zc4h2 | -0.059 |
| smim29 | 0.114 | scrt2 | -0.058 |
| rabl6b | 0.114 | marcksb | -0.058 |
| klf6a | 0.113 | ptmab | -0.057 |
| tuba8l3 | 0.112 | pbx3b | -0.056 |
| ctsd | 0.110 | hbae3 | -0.056 |
| degs1 | 0.110 | gpr85 | -0.056 |
| slc37a2 | 0.108 | cadm3 | -0.055 |
| eno3 | 0.106 | elavl4 | -0.055 |
| npc2 | 0.105 | actc1b | -0.055 |
| zgc:153867 | 0.104 | golga7ba | -0.054 |
| idh1 | 0.104 | myt1b | -0.054 |
| fxyd1 | 0.103 | vamp2 | -0.054 |
| anxa11b | 0.103 | dlb | -0.053 |
| elovl1b | 0.103 | tuba2 | -0.053 |
| lgals3a | 0.103 | mylpfa | -0.053 |
| osbpl10b | 0.102 | cbfa2t3 | -0.052 |
| capns1a | 0.102 | hmgb1b | -0.052 |
| sypl2b | 0.101 | fabp7a | -0.052 |
| chmp5b | 0.101 | id4 | -0.052 |
| myl12.1 | 0.100 | hbbe2 | -0.052 |