"rhomboid, veinlet-like 3 (Drosophila)"
ZFIN 
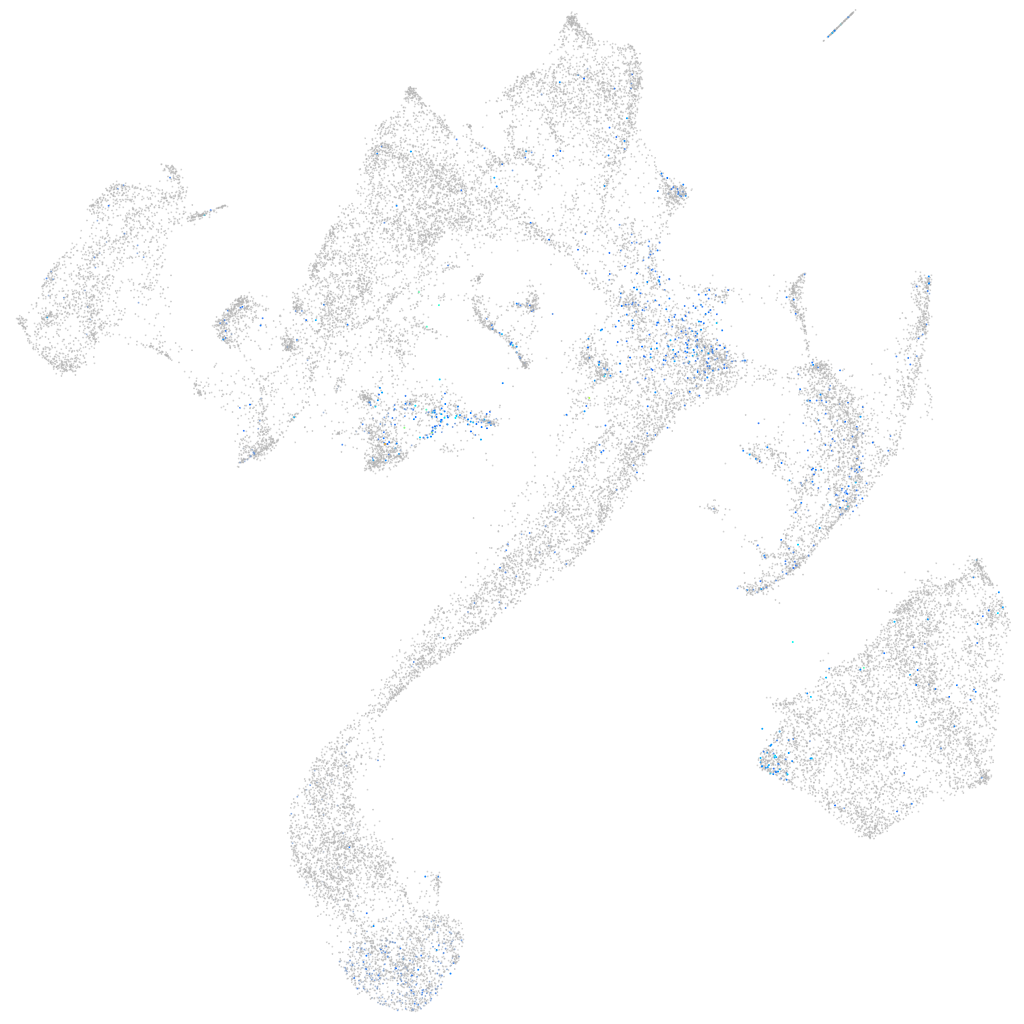
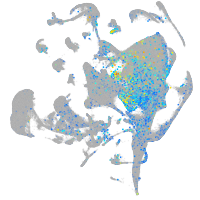
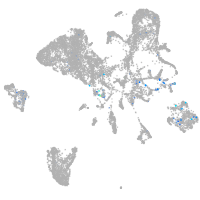
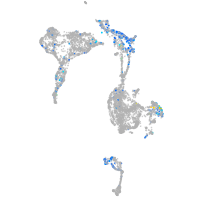
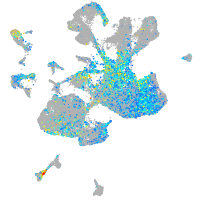
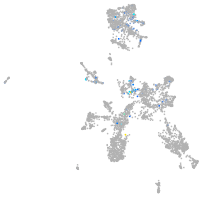
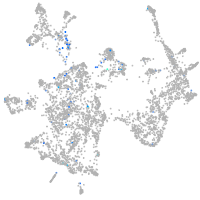
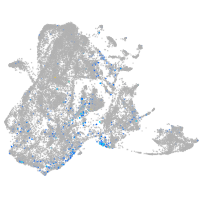
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-265d7.2 | 0.122 | fxr2 | -0.046 |
| LOC798783 | 0.118 | CABZ01072309.1 | -0.044 |
| XLOC-003689 | 0.110 | plecb | -0.044 |
| gadd45gb.1 | 0.099 | si:dkey-74k8.3 | -0.043 |
| abhd6a | 0.093 | eef1da | -0.043 |
| si:ch73-21g5.7 | 0.092 | nid1a | -0.043 |
| vim | 0.090 | col1a1a | -0.042 |
| CR812832.1 | 0.090 | col1a2 | -0.042 |
| insm1a | 0.089 | actc1a | -0.040 |
| zgc:165461 | 0.089 | desma | -0.040 |
| XLOC-003692 | 0.088 | cox17 | -0.040 |
| elavl3 | 0.085 | CABZ01072309.2 | -0.038 |
| XLOC-003690 | 0.085 | srl | -0.038 |
| prss60.2 | 0.084 | fitm1 | -0.038 |
| ldlrad2 | 0.084 | cox7c | -0.038 |
| neurod4 | 0.080 | tnnt2c | -0.037 |
| dlb | 0.080 | casq2 | -0.037 |
| scrt2 | 0.079 | rps18 | -0.037 |
| nhlh2 | 0.079 | acta1a | -0.036 |
| myt1b | 0.076 | rpl29 | -0.036 |
| cxcr4a | 0.076 | pabpc4 | -0.035 |
| her4.4 | 0.075 | bin1b | -0.035 |
| her4.2 | 0.075 | atp2a1 | -0.035 |
| gfap | 0.075 | tnn | -0.035 |
| CU634008.1 | 0.074 | rbfox1l | -0.034 |
| CSDC2 | 0.074 | serpinf1 | -0.034 |
| cblc | 0.074 | cdh15 | -0.034 |
| mir219-3 | 0.073 | tnni2b.1 | -0.034 |
| msi1 | 0.071 | tnnc1b | -0.034 |
| her13 | 0.070 | mef2aa | -0.034 |
| nkx2.2b | 0.069 | col1a1b | -0.033 |
| fn1b | 0.069 | sgcd | -0.033 |
| phox2bb | 0.068 | fabp3 | -0.033 |
| insm1b | 0.068 | zgc:101853 | -0.033 |
| pou3f1 | 0.068 | klhl31 | -0.033 |