regulator of G protein signaling 6
ZFIN 
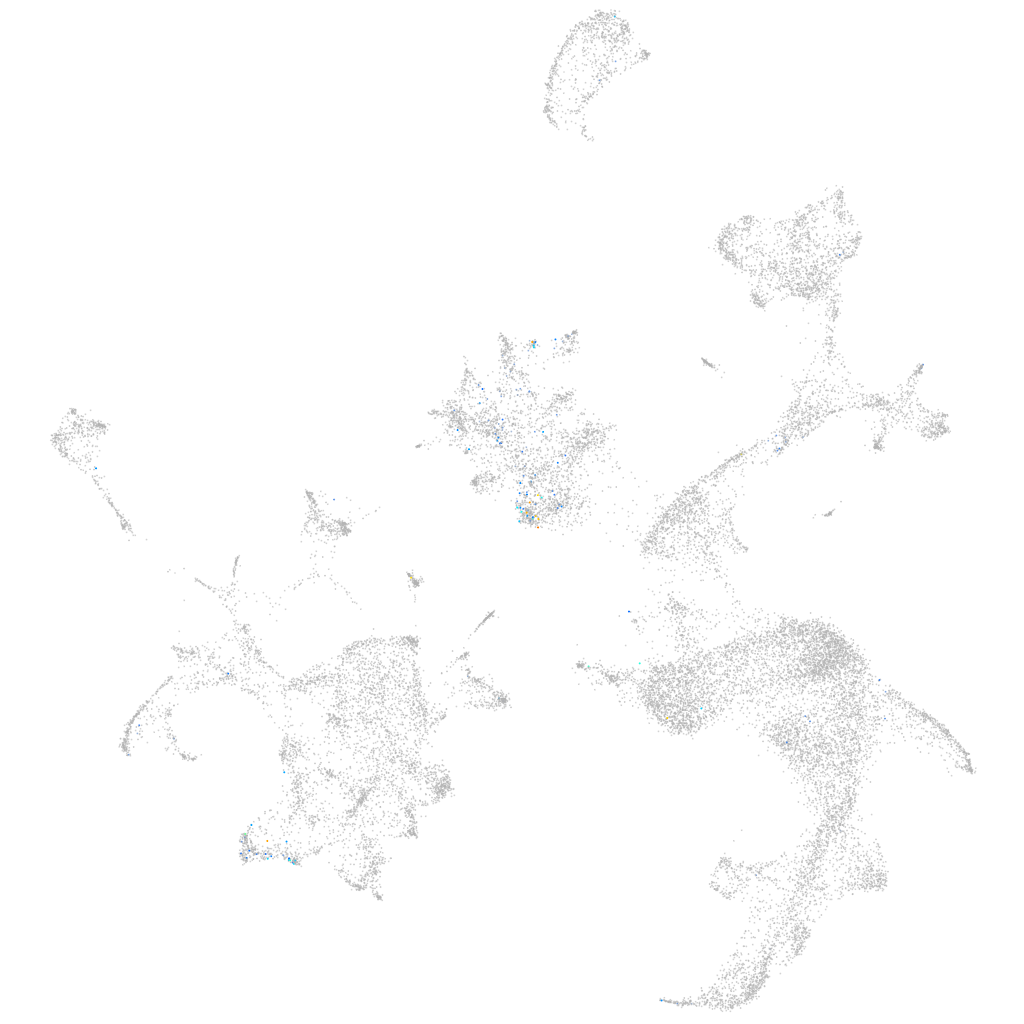
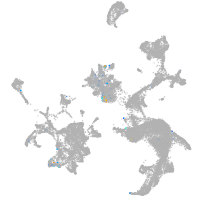
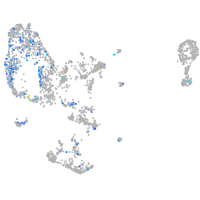
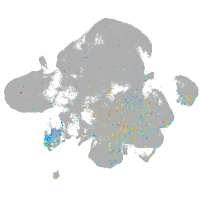
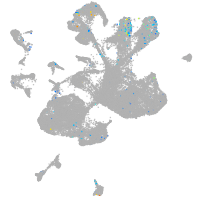
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-33m11.7 | 0.196 | hbbe3 | -0.027 |
| cldn23b | 0.187 | slc11a2 | -0.026 |
| kcna1b | 0.168 | fam210b | -0.022 |
| BX530079.1 | 0.167 | hmbsb | -0.022 |
| kcnj9 | 0.165 | urod | -0.022 |
| chrm4a | 0.150 | gp1bb | -0.021 |
| CR376751.1 | 0.149 | slc4a1a | -0.021 |
| rom1a | 0.145 | alad | -0.021 |
| BX908756.2 | 0.140 | anp32b | -0.020 |
| unc119.2 | 0.137 | fech | -0.019 |
| prokr1b | 0.136 | rab11fip1b | -0.019 |
| ptprna | 0.133 | si:dkey-93h22.7 | -0.019 |
| gabra1 | 0.132 | hmbsa | -0.019 |
| edil3a | 0.131 | ctsz | -0.018 |
| si:dkey-22h13.5 | 0.130 | blvrb | -0.018 |
| XLOC-019901 | 0.130 | zgc:56699 | -0.017 |
| nmnat2 | 0.128 | rnaseka | -0.017 |
| CR589947.2 | 0.128 | blvra | -0.017 |
| saga | 0.125 | plac8l1 | -0.017 |
| vamp2 | 0.125 | rhd | -0.017 |
| nrl | 0.124 | tmem14ca | -0.017 |
| mir132-2 | 0.121 | uros | -0.017 |
| spa17 | 0.120 | tspo | -0.016 |
| kif3cb | 0.119 | coro1a | -0.016 |
| adgra1a | 0.116 | fam53b | -0.016 |
| disp2 | 0.116 | dnase1l4.2 | -0.016 |
| eno2 | 0.116 | arl4aa | -0.016 |
| LOC103911284 | 0.116 | ell2 | -0.016 |
| sagb | 0.114 | zgc:163057 | -0.016 |
| guca1b | 0.113 | mafbb | -0.016 |
| si:ch211-113d22.2 | 0.112 | cldng | -0.016 |
| cnga1b | 0.112 | ppox | -0.016 |
| si:ch211-193i15.2 | 0.112 | si:ch73-248e21.7 | -0.015 |
| grm4 | 0.112 | hdr | -0.015 |
| BX004774.2 | 0.111 | si:ch211-114n24.6 | -0.015 |