regulator of G protein signaling 6
ZFIN 
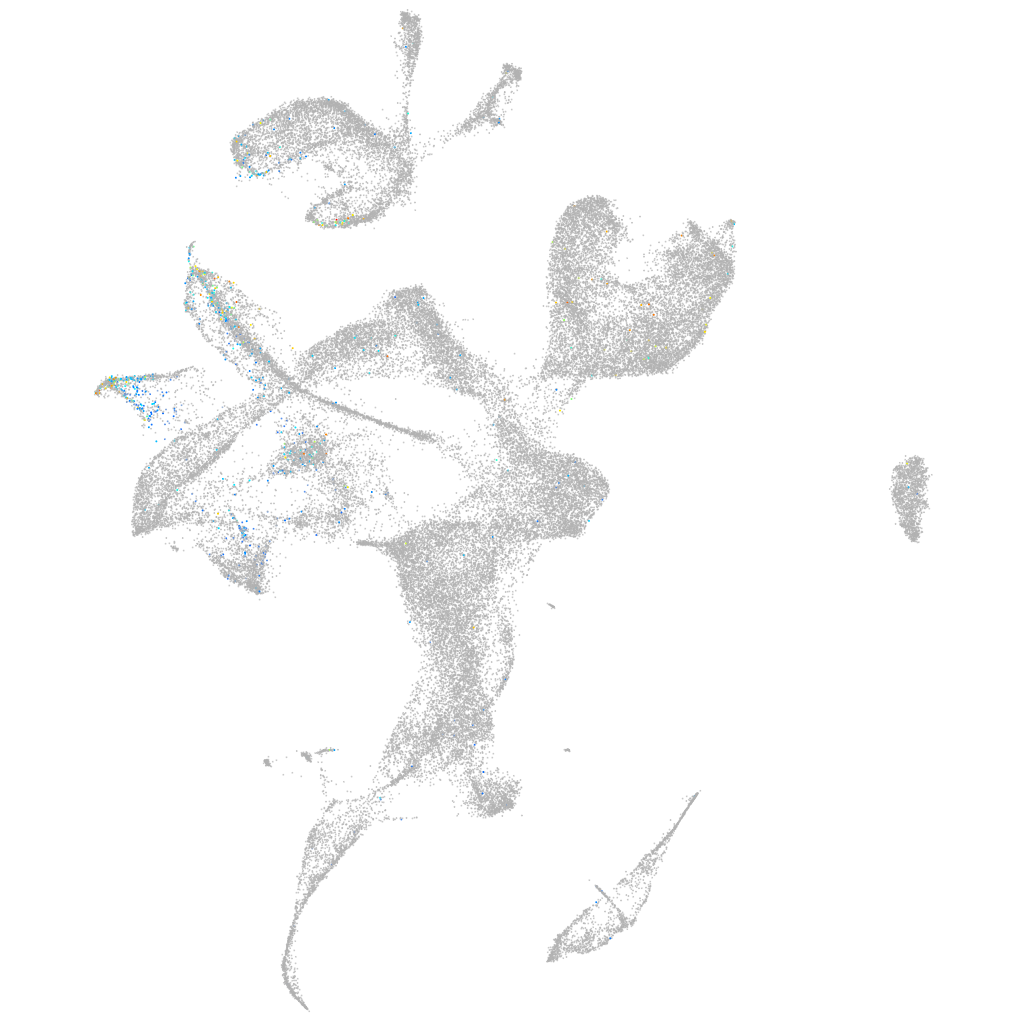
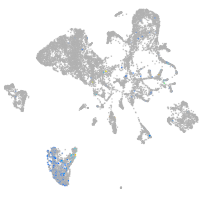
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pde6b | 0.249 | hmga1a | -0.075 |
| grk1a | 0.243 | hmgb2a | -0.072 |
| rom1a | 0.237 | hmgb2b | -0.067 |
| pde6a | 0.231 | hmgn2 | -0.063 |
| rom1b | 0.228 | h2afvb | -0.060 |
| sagb | 0.228 | si:ch211-222l21.1 | -0.059 |
| saga | 0.226 | si:dkey-151g10.6 | -0.059 |
| gnat1 | 0.225 | rplp0 | -0.059 |
| rhol | 0.217 | rplp1 | -0.057 |
| pdca | 0.216 | rps12 | -0.056 |
| si:ch211-113d22.2 | 0.216 | rpl9 | -0.056 |
| rho | 0.214 | rps23 | -0.056 |
| gngt1 | 0.212 | ahcy | -0.056 |
| cnga1b | 0.212 | tuba8l4 | -0.055 |
| cplx4c | 0.211 | snrpf | -0.055 |
| guca1b | 0.207 | rpl11 | -0.055 |
| BX004774.2 | 0.205 | rpl12 | -0.055 |
| CABZ01084566.1 | 0.204 | pcna | -0.055 |
| rbp2a | 0.201 | rps9 | -0.054 |
| cnga1a | 0.195 | rps13 | -0.054 |
| gucy2f | 0.184 | rpl10 | -0.054 |
| pde6ga | 0.182 | rrm1 | -0.053 |
| zgc:112334 | 0.176 | naca | -0.053 |
| zgc:162144 | 0.167 | rpl27 | -0.052 |
| guca1a | 0.163 | rpl8 | -0.052 |
| aipl1 | 0.152 | rplp2l | -0.052 |
| si:ch211-207j7.2 | 0.151 | tpt1 | -0.052 |
| cngb1a | 0.149 | stmn1a | -0.052 |
| cabp4 | 0.143 | mdka | -0.052 |
| zgc:77752 | 0.140 | rpl27a | -0.051 |
| gc2 | 0.137 | rps19 | -0.051 |
| unc119.2 | 0.136 | ran | -0.051 |
| eno2 | 0.135 | snrpd1 | -0.051 |
| gnb1a | 0.135 | rps2 | -0.051 |
| LOC101884106 | 0.134 | anp32b | -0.051 |