repulsive guidance molecule BMP co-receptor a
ZFIN 
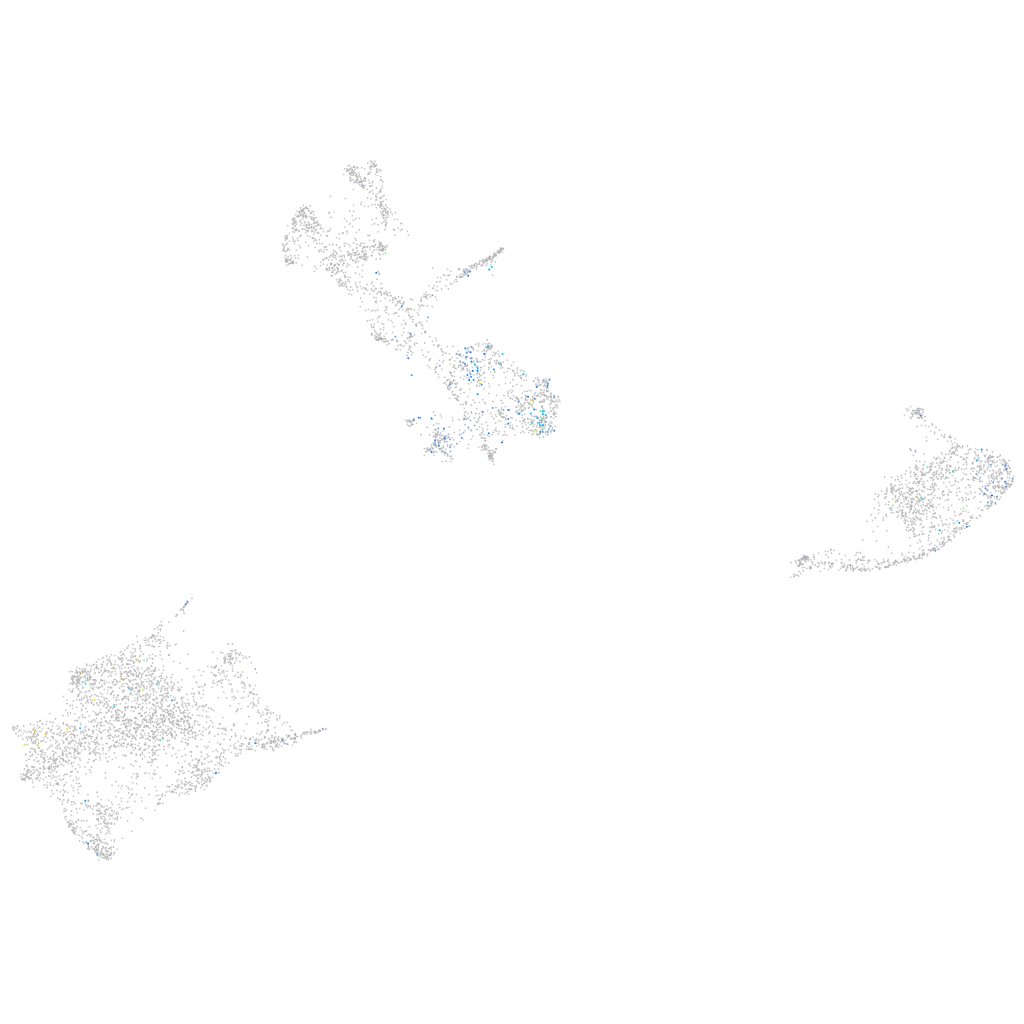
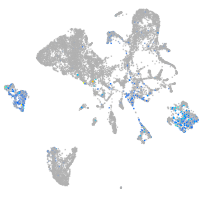
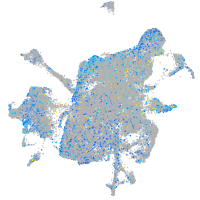
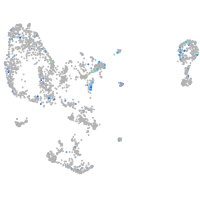
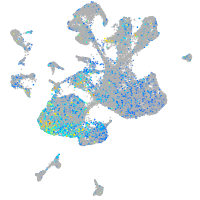
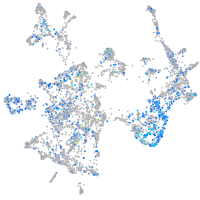
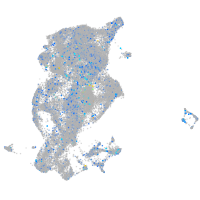
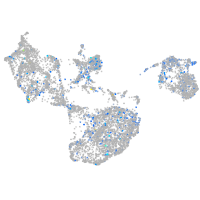
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prlh2 | 0.166 | slc2a15a | -0.065 |
| prok1 | 0.160 | prps1a | -0.060 |
| CABZ01011209.1 | 0.159 | agtrap | -0.059 |
| CABZ01052570.1 | 0.152 | gmps | -0.059 |
| XLOC-032440 | 0.151 | pts | -0.058 |
| gng3 | 0.148 | paics | -0.057 |
| phf21aa | 0.145 | glulb | -0.057 |
| zgc:198371 | 0.145 | impdh1b | -0.055 |
| nova2 | 0.143 | si:dkey-151g10.6 | -0.051 |
| fev | 0.142 | atic | -0.049 |
| cpne9 | 0.137 | gstp1 | -0.049 |
| zgc:174564 | 0.136 | prdx1 | -0.048 |
| gpm6aa | 0.135 | rplp1 | -0.047 |
| tuba1c | 0.134 | zgc:165573 | -0.047 |
| syt9a | 0.131 | pnp4a | -0.046 |
| ptprn2 | 0.131 | akr1b1 | -0.046 |
| gabrb4 | 0.130 | trpm1b | -0.045 |
| elavl3 | 0.129 | ppat | -0.045 |
| vamp2 | 0.129 | inka1a | -0.045 |
| rtn1a | 0.129 | rab32a | -0.045 |
| mras | 0.128 | cax1 | -0.044 |
| CABZ01043955.1 | 0.128 | ponzr1 | -0.044 |
| LO017650.1 | 0.128 | si:dkey-197i20.6 | -0.043 |
| tph2 | 0.127 | sigmar1 | -0.043 |
| rapgefl1 | 0.127 | zgc:110789 | -0.043 |
| actn2a | 0.126 | slc2a11b | -0.043 |
| syt1a | 0.126 | tagln3b | -0.043 |
| foxj1b | 0.126 | shmt1 | -0.042 |
| xpr1a | 0.126 | anxa4 | -0.042 |
| tspan13b | 0.124 | shmt2 | -0.042 |
| sncb | 0.124 | cd164 | -0.042 |
| grpr | 0.123 | si:ch211-210c8.7 | -0.042 |
| kif1ab | 0.122 | rnaset2 | -0.042 |
| kcnc4 | 0.122 | bco1 | -0.041 |
| tuba2 | 0.121 | ctsba | -0.041 |