"REX1, RNA exonuclease 1 homolog"
ZFIN 
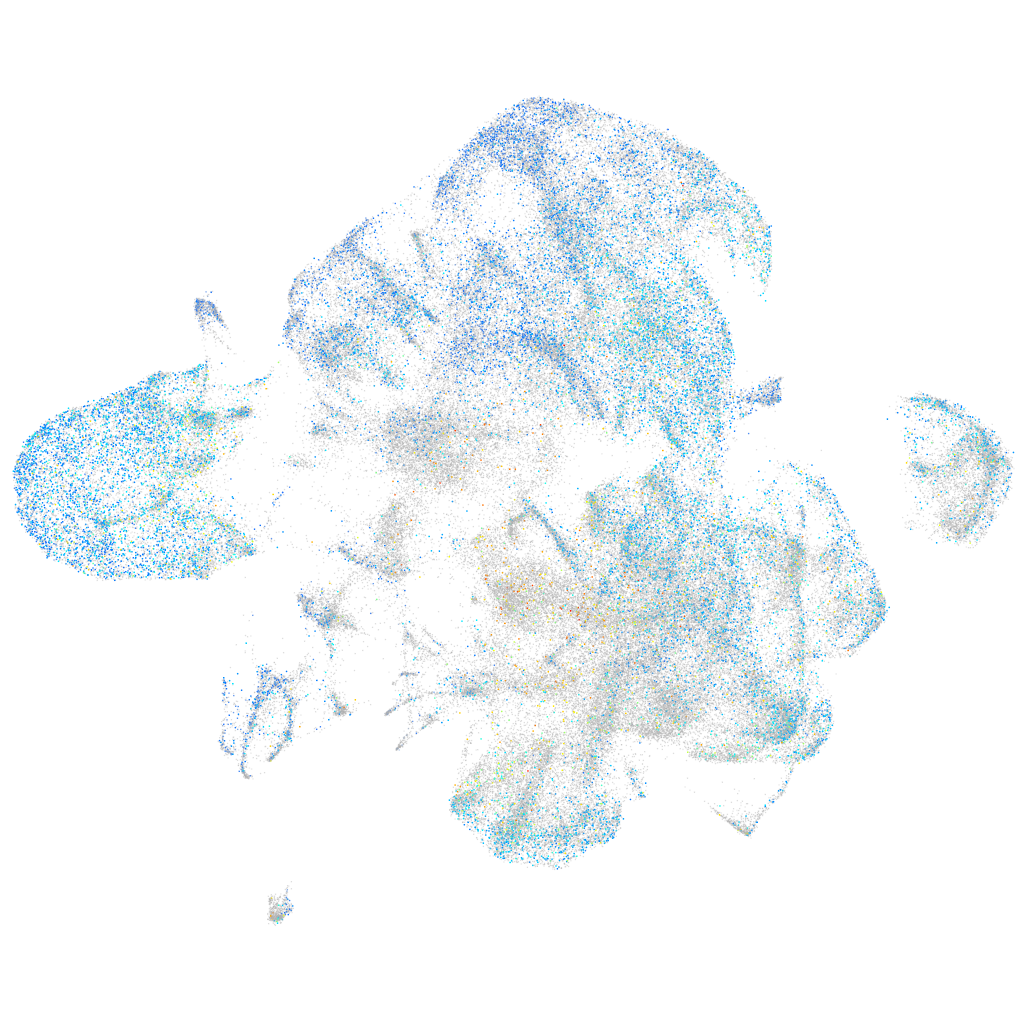
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpub | 0.126 | rpl37 | -0.084 |
| si:ch211-152c2.3 | 0.125 | rps10 | -0.075 |
| stm | 0.121 | zgc:114188 | -0.068 |
| NC-002333.4 | 0.118 | gapdhs | -0.067 |
| hspb1 | 0.114 | ptmaa | -0.065 |
| top1l | 0.113 | ppiab | -0.063 |
| hnrnpa1b | 0.111 | atp6v0cb | -0.062 |
| pou5f3 | 0.109 | h3f3a | -0.061 |
| rbm4.3 | 0.108 | cspg5a | -0.058 |
| apoeb | 0.107 | tpi1b | -0.057 |
| ilf3b | 0.104 | calm1a | -0.057 |
| khdrbs1a | 0.104 | ywhag2 | -0.056 |
| crabp2b | 0.104 | sncb | -0.056 |
| hnrnpm | 0.103 | pvalb1 | -0.055 |
| polr3gla | 0.103 | actc1b | -0.055 |
| s100a1 | 0.103 | calm1b | -0.054 |
| syncrip | 0.102 | sncgb | -0.053 |
| hmga1a | 0.102 | pvalb2 | -0.052 |
| seta | 0.102 | sv2a | -0.052 |
| si:dkey-66i24.9 | 0.102 | sypb | -0.051 |
| cx43.4 | 0.102 | snap25a | -0.051 |
| smarca4a | 0.100 | atp6ap2 | -0.051 |
| snrnp70 | 0.099 | zgc:65894 | -0.051 |
| bms1 | 0.098 | eno1a | -0.050 |
| hnrnpabb | 0.098 | zgc:158463 | -0.050 |
| anp32e | 0.098 | stmn2a | -0.049 |
| rtca | 0.098 | slc6a1a | -0.048 |
| zmp:0000000624 | 0.098 | stxbp1a | -0.048 |
| COX7A2 | 0.097 | nptna | -0.048 |
| apela | 0.097 | atpv0e2 | -0.048 |
| safb | 0.096 | scg2b | -0.048 |
| si:dkey-56m19.5 | 0.096 | aldocb | -0.048 |
| hnrnpa1a | 0.096 | apoa2 | -0.047 |
| apoc1 | 0.095 | ndufa4 | -0.047 |
| sf3b2 | 0.094 | calm3a | -0.047 |