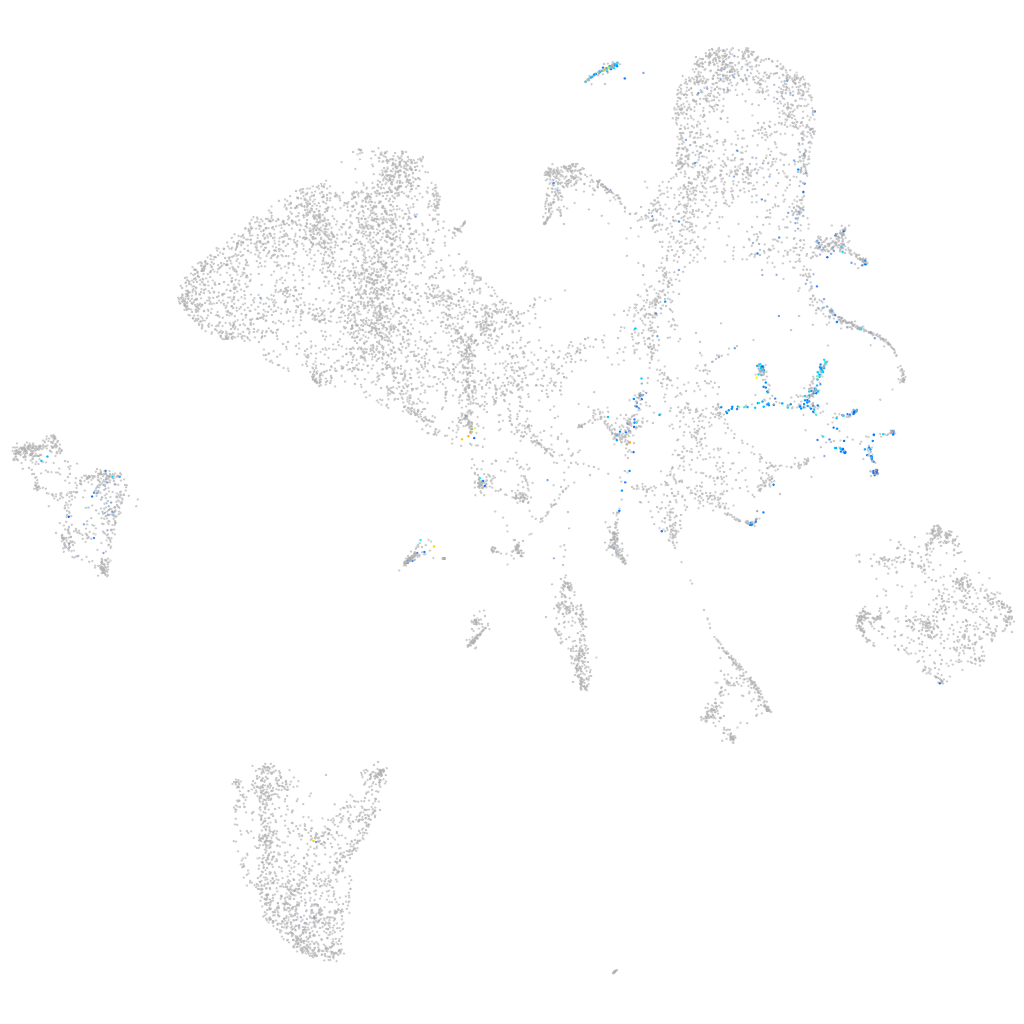
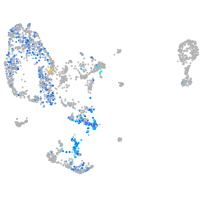
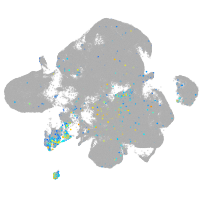
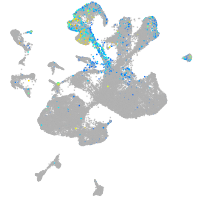
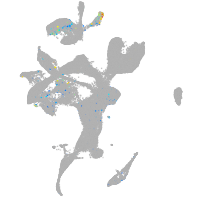
ret proto-oncogene receptor tyrosine kinase
ZFIN 
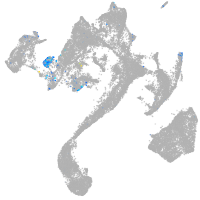
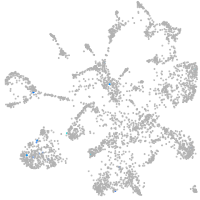
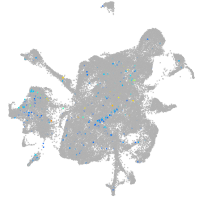
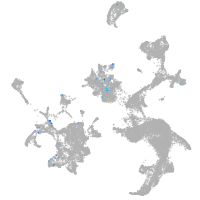
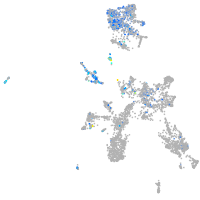
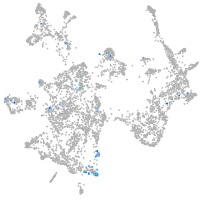
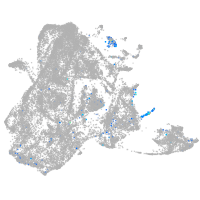
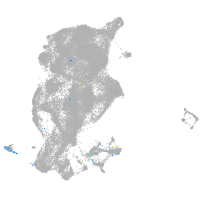
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.496 | gamt | -0.156 |
| insm1b | 0.449 | ahcy | -0.145 |
| neurod1 | 0.440 | gatm | -0.144 |
| mir7a-1 | 0.377 | bhmt | -0.143 |
| zgc:101731 | 0.377 | gapdh | -0.142 |
| nkx2.2a | 0.372 | mat1a | -0.142 |
| pcsk1nl | 0.372 | fabp3 | -0.134 |
| fev | 0.369 | aqp12 | -0.130 |
| pax6b | 0.359 | apoc1 | -0.128 |
| hepacam2 | 0.355 | apoa1b | -0.127 |
| dll4 | 0.355 | cx32.3 | -0.124 |
| mir375-2 | 0.351 | apoa4b.1 | -0.124 |
| c2cd4a | 0.350 | agxtb | -0.123 |
| elovl1a | 0.337 | fbp1b | -0.123 |
| scgn | 0.334 | nupr1b | -0.122 |
| id4 | 0.332 | aldob | -0.122 |
| egr4 | 0.330 | si:dkey-16p21.8 | -0.121 |
| insm1a | 0.329 | aldh6a1 | -0.121 |
| syt1a | 0.326 | hdlbpa | -0.120 |
| pcsk1 | 0.321 | apoc2 | -0.119 |
| myt1b | 0.309 | aldh7a1 | -0.119 |
| rims2a | 0.308 | agxta | -0.118 |
| vat1 | 0.303 | mgst1.2 | -0.118 |
| hpcal1 | 0.302 | afp4 | -0.118 |
| vamp2 | 0.300 | adka | -0.117 |
| si:ch73-160i9.2 | 0.299 | tfa | -0.116 |
| lysmd2 | 0.298 | serpina1 | -0.115 |
| scg2a | 0.298 | apoa2 | -0.114 |
| trmt9b | 0.291 | wu:fj16a03 | -0.114 |
| tox | 0.289 | gnmt | -0.113 |
| capsla | 0.288 | ttc36 | -0.113 |
| ptprna | 0.283 | shmt1 | -0.113 |
| chga | 0.279 | serpina1l | -0.113 |
| zgc:165481 | 0.277 | ftcd | -0.111 |
| calm1b | 0.277 | ambp | -0.110 |