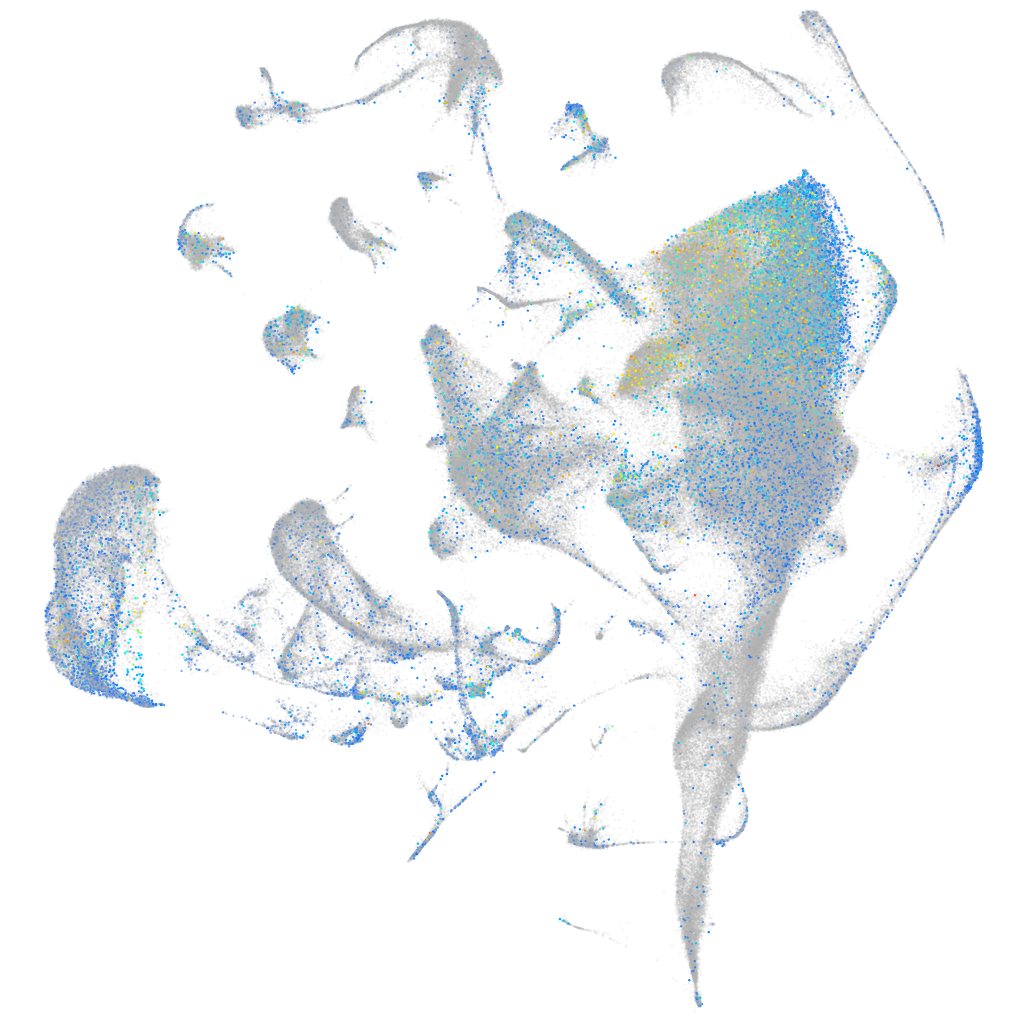"RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing"
ZFIN 
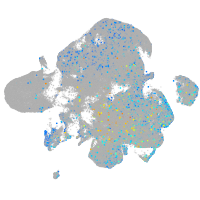
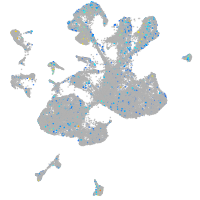
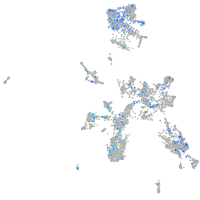
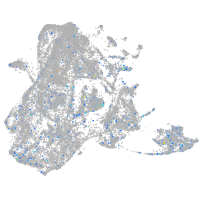
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.094 | hspb1 | -0.052 |
| zgc:65894 | 0.094 | si:ch211-152c2.3 | -0.052 |
| gng3 | 0.093 | pou5f3 | -0.050 |
| stmn2a | 0.093 | stm | -0.050 |
| ywhag2 | 0.091 | apoc1 | -0.047 |
| vamp2 | 0.090 | dkc1 | -0.046 |
| elavl4 | 0.089 | si:dkey-66i24.9 | -0.046 |
| mllt11 | 0.088 | apoeb | -0.045 |
| rtn1b | 0.088 | npm1a | -0.045 |
| stxbp1a | 0.088 | polr3gla | -0.045 |
| stx1b | 0.086 | fbl | -0.044 |
| tuba1c | 0.086 | nop58 | -0.044 |
| stmn1b | 0.085 | s100a1 | -0.044 |
| ywhah | 0.084 | tbx16 | -0.044 |
| gap43 | 0.082 | si:ch1073-80i24.3 | -0.043 |
| gnao1a | 0.082 | vox | -0.043 |
| snap25a | 0.082 | zgc:56699 | -0.043 |
| atp6v0cb | 0.081 | akap12b | -0.042 |
| gnb1a | 0.081 | asb11 | -0.042 |
| atp6v1e1b | 0.080 | cdca7a | -0.042 |
| rnasekb | 0.080 | nnr | -0.042 |
| atp6v1g1 | 0.079 | ved | -0.042 |
| gdi1 | 0.079 | zmp:0000000624 | -0.042 |
| elavl3 | 0.078 | COX7A2 | -0.041 |
| gng2 | 0.078 | ddx18 | -0.041 |
| calm1a | 0.078 | gar1 | -0.041 |
| map1aa | 0.077 | ing5b | -0.041 |
| zgc:153426 | 0.077 | lig1 | -0.041 |
| cnrip1a | 0.076 | bms1 | -0.040 |
| maptb | 0.076 | cx43.4 | -0.040 |
| rtn1a | 0.076 | lrwd1 | -0.040 |
| sv2a | 0.075 | mki67 | -0.039 |
| fez1 | 0.074 | nop56 | -0.039 |
| si:dkeyp-75h12.5 | 0.074 | rsl1d1 | -0.039 |
| tubb5 | 0.074 | ccnb1 | -0.038 |