RecQ helicase-like 4
ZFIN 
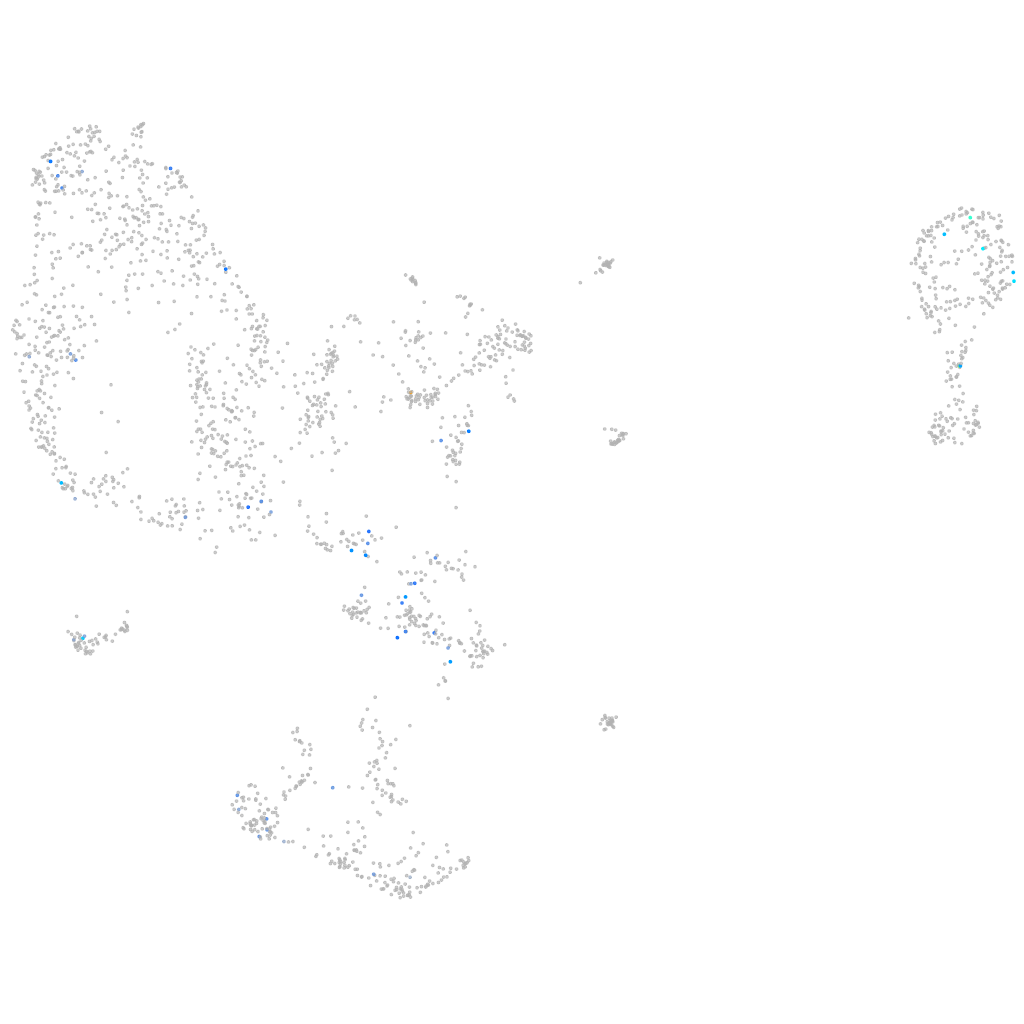
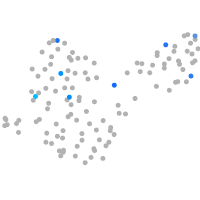
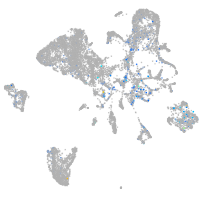
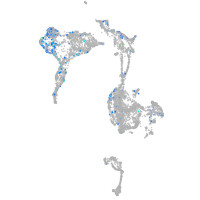
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| urgcp | 0.396 | rpl10 | -0.086 |
| aplnr2 | 0.323 | eno3 | -0.083 |
| sgsm1a | 0.316 | aldob | -0.080 |
| CABZ01063543.1 | 0.302 | dap1b | -0.077 |
| si:dkey-149m13.4 | 0.302 | atp5meb | -0.075 |
| si:ch211-212k18.5 | 0.295 | rplp2 | -0.074 |
| XLOC-011414 | 0.292 | rps26 | -0.074 |
| zgc:113223 | 0.274 | atp5l | -0.073 |
| zgc:173856 | 0.261 | ctsla | -0.073 |
| CABZ01048956.1 | 0.254 | cox6c | -0.073 |
| CABZ01043955.1 | 0.253 | atp5mc3b | -0.073 |
| si:dkey-185m8.2 | 0.250 | glrx | -0.072 |
| trim36 | 0.245 | eef1da | -0.071 |
| lyplal1 | 0.244 | tpi1b | -0.070 |
| asf1ba | 0.238 | nit2 | -0.070 |
| FP016056.1 | 0.228 | eif4a1b | -0.069 |
| pabpc1l | 0.226 | gapdh | -0.069 |
| mrc2 | 0.226 | rpl9 | -0.069 |
| si:ch211-145h19.5 | 0.222 | stau2 | -0.068 |
| ubac2 | 0.221 | atp5f1d | -0.068 |
| CU179702.2 | 0.218 | gcshb | -0.068 |
| asz1 | 0.214 | gamt | -0.067 |
| BX663613.1 | 0.211 | fbp1b | -0.067 |
| rab34b | 0.211 | grhprb | -0.067 |
| CR339054.1 | 0.208 | elovl1b | -0.067 |
| XLOC-013311 | 0.208 | romo1 | -0.067 |
| CR547131.1 | 0.207 | tob1b | -0.066 |
| zgc:110540 | 0.204 | cox7c | -0.065 |
| LOC103911090 | 0.200 | npc2 | -0.065 |
| BX324206.1 | 0.199 | gstp1 | -0.065 |
| pimr193 | 0.198 | adh8b | -0.065 |
| serp2 | 0.197 | actb2 | -0.064 |
| nlrc5 | 0.194 | alpl | -0.064 |
| XLOC-029591 | 0.194 | zgc:101040 | -0.064 |
| BX890600.1 | 0.191 | gabarapa | -0.064 |