reversion-inducing-cysteine-rich protein with kazal motifs
ZFIN 
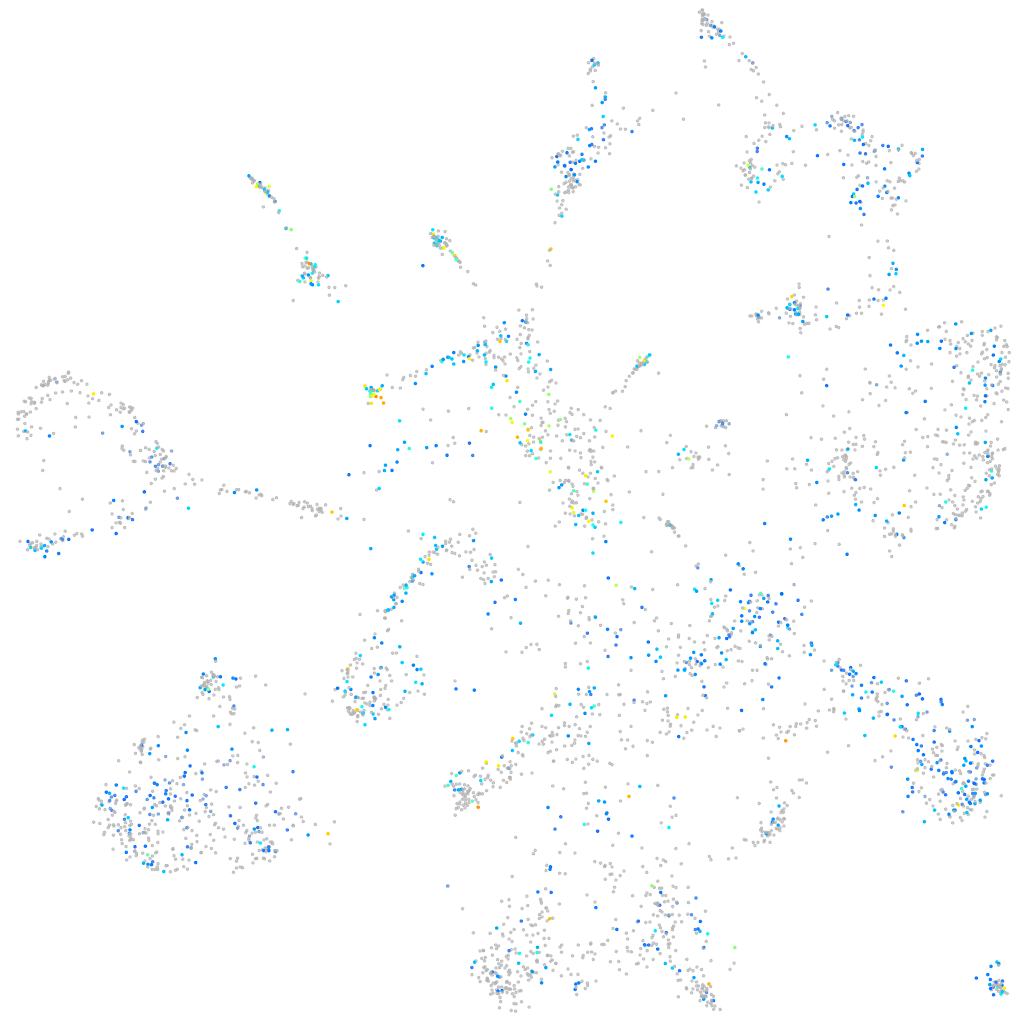
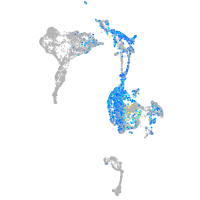
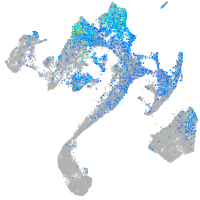
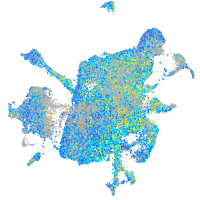
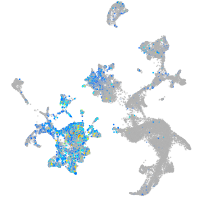
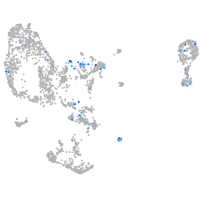
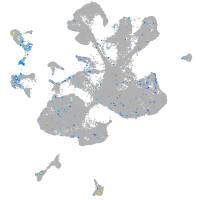
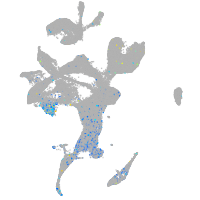
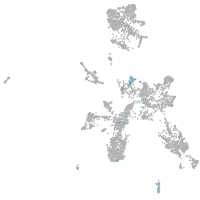
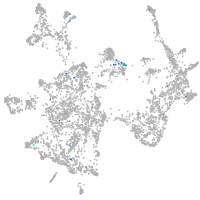
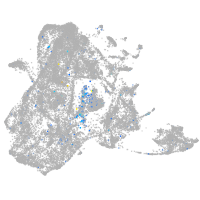
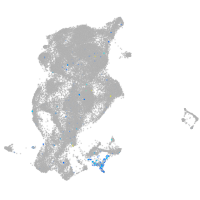
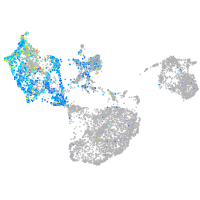
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnn | 0.189 | csrp1b | -0.113 |
| lamc3 | 0.161 | cald1b | -0.100 |
| fmoda | 0.158 | krt15 | -0.099 |
| si:dkeyp-106c3.1 | 0.158 | desmb | -0.099 |
| matn4 | 0.152 | cavin2b | -0.089 |
| col2a1b | 0.150 | acta2 | -0.083 |
| pdgfrl | 0.149 | fn1b | -0.083 |
| cnmd | 0.149 | cxcl14 | -0.079 |
| slc6a4a | 0.148 | rbm24a | -0.078 |
| pdgfrb | 0.145 | arhgap20b | -0.076 |
| foxc1a | 0.139 | vcla | -0.076 |
| dcn | 0.137 | ckbb | -0.074 |
| foxa1 | 0.137 | myl6 | -0.073 |
| ednraa | 0.137 | akap7 | -0.073 |
| wfdc2 | 0.136 | rgs2 | -0.072 |
| col5a1 | 0.135 | myl9a | -0.072 |
| fbn2b | 0.130 | zgc:110699 | -0.072 |
| col11a1b | 0.129 | cnn1b | -0.071 |
| itga11a | 0.128 | itpka | -0.070 |
| si:ch211-106j24.1 | 0.127 | tpm2 | -0.070 |
| notch3 | 0.126 | XLOC-041870 | -0.069 |
| nid1b | 0.124 | pgam1a | -0.069 |
| foxc1b | 0.123 | cavin1b | -0.068 |
| cald1a | 0.122 | myo1ca | -0.068 |
| twist2 | 0.122 | fhl1a | -0.068 |
| pmp22a | 0.117 | si:ch211-137i24.10 | -0.067 |
| zeb2a | 0.116 | pdlim3a | -0.067 |
| hapln1a | 0.116 | fbp2 | -0.067 |
| ebf1a | 0.115 | fbxl22 | -0.066 |
| cdh11 | 0.114 | eno1a | -0.066 |
| sept2 | 0.114 | krt91 | -0.065 |
| si:dkey-238c7.16 | 0.114 | si:dkeyp-66d1.7 | -0.065 |
| cd248a | 0.113 | pdlim3b | -0.064 |
| SCARF2 | 0.113 | slc16a3 | -0.064 |
| nkx3.2 | 0.113 | pkma | -0.064 |