rasd family member 4
ZFIN 
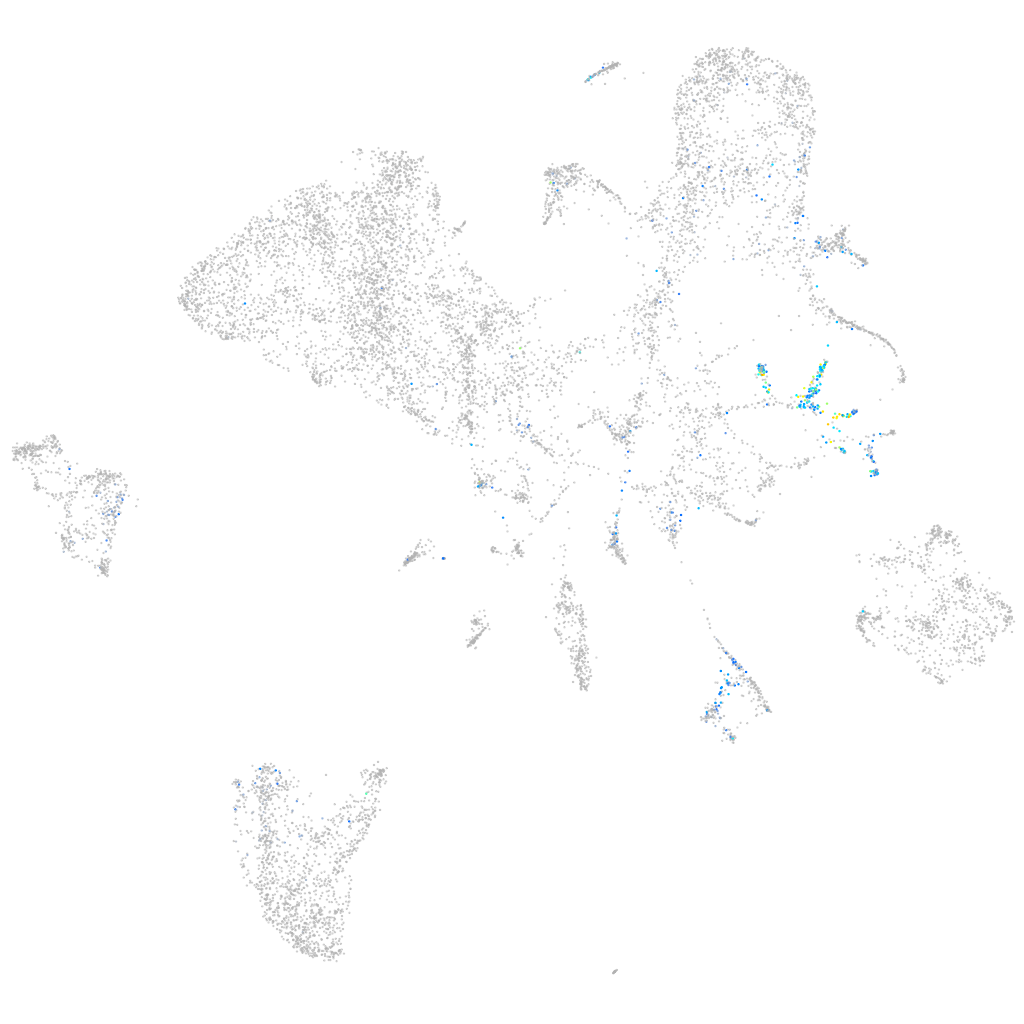
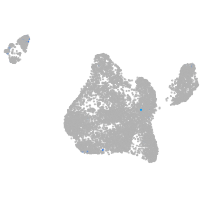
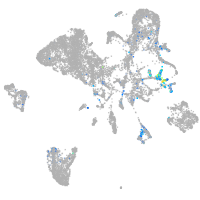
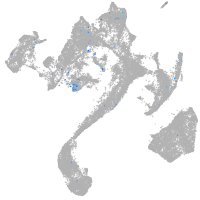
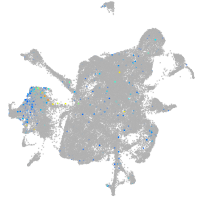
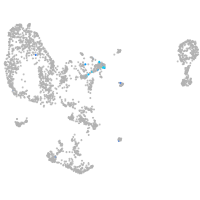
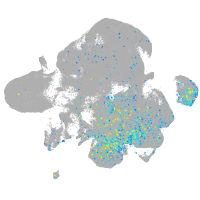
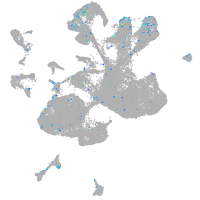
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.556 | ahcy | -0.162 |
| scg3 | 0.502 | gapdh | -0.159 |
| arxa | 0.481 | gamt | -0.157 |
| neurod1 | 0.468 | gatm | -0.147 |
| pax6b | 0.455 | bhmt | -0.144 |
| si:dkey-153k10.9 | 0.440 | fabp3 | -0.135 |
| pcsk2 | 0.431 | mat1a | -0.134 |
| syt1a | 0.423 | apoa1b | -0.134 |
| egr4 | 0.423 | hdlbpa | -0.132 |
| rprmb | 0.412 | apoc1 | -0.132 |
| ccka | 0.403 | apoa4b.1 | -0.131 |
| pcsk1nl | 0.400 | apoc2 | -0.130 |
| c2cd4a | 0.400 | afp4 | -0.126 |
| gcga | 0.397 | fbp1b | -0.124 |
| vat1 | 0.395 | aldh6a1 | -0.123 |
| kcnj11 | 0.393 | mgst1.2 | -0.122 |
| mir7a-1 | 0.382 | gstt1a | -0.122 |
| tspan7b | 0.379 | si:dkey-16p21.8 | -0.121 |
| LOC100537384 | 0.376 | agxtb | -0.121 |
| fev | 0.374 | cx32.3 | -0.119 |
| isl1 | 0.373 | apoa2 | -0.118 |
| abhd15a | 0.364 | agxta | -0.118 |
| rem2 | 0.358 | gstr | -0.118 |
| insm1b | 0.354 | aqp12 | -0.117 |
| hepacam2 | 0.352 | grhprb | -0.116 |
| pcsk1 | 0.349 | aldh7a1 | -0.116 |
| mir375-2 | 0.346 | apobb.1 | -0.116 |
| calm1b | 0.346 | glud1b | -0.115 |
| nkx2.2a | 0.345 | scp2a | -0.115 |
| si:ch73-160i9.2 | 0.337 | gnmt | -0.115 |
| kcnk9 | 0.335 | serpina1 | -0.114 |
| capsla | 0.332 | aldob | -0.113 |
| scg2a | 0.330 | serpina1l | -0.112 |
| etv1 | 0.328 | apom | -0.112 |
| gfra3 | 0.324 | fetub | -0.111 |