RAD51 associated protein 1
ZFIN 
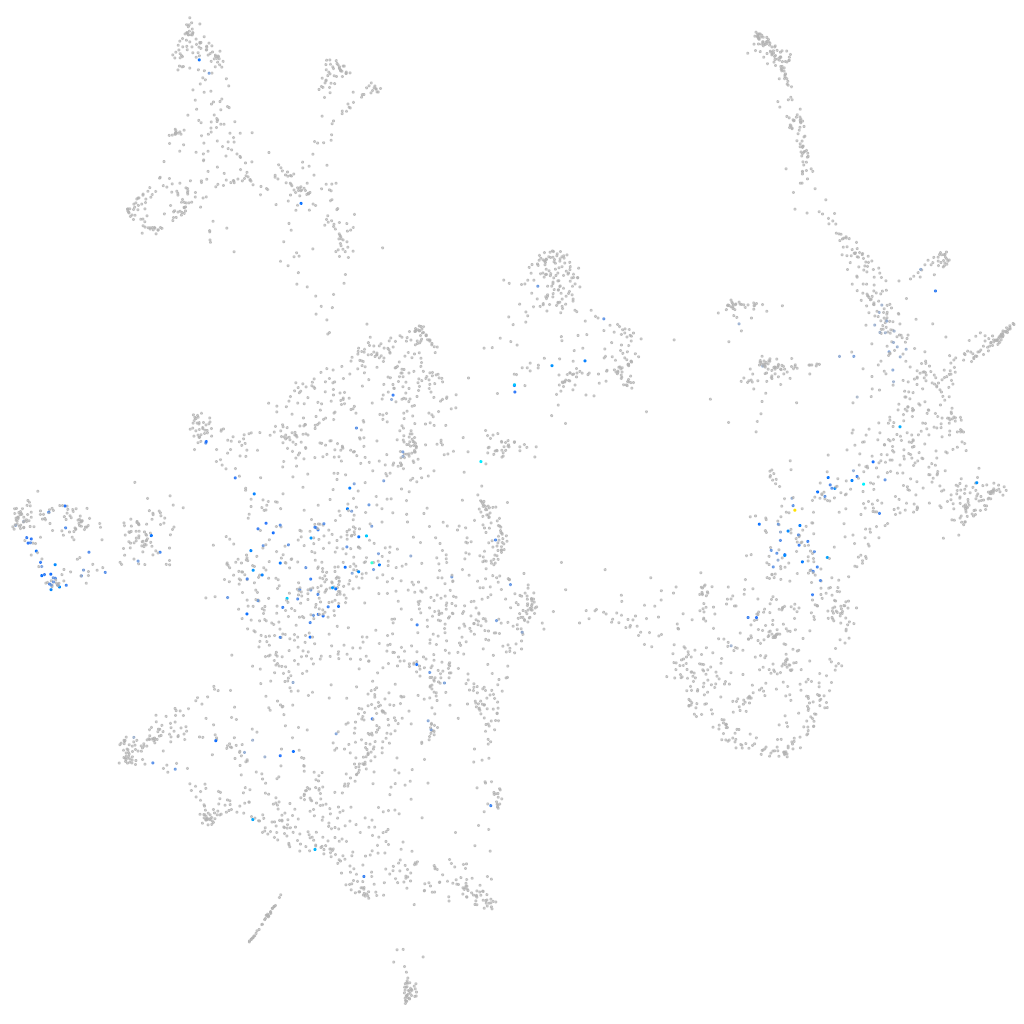
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110216 | 0.279 | eno1a | -0.076 |
| si:ch211-113a14.18 | 0.278 | mt-co2 | -0.075 |
| ccna2 | 0.276 | CR383676.1 | -0.075 |
| mibp | 0.268 | tob1b | -0.075 |
| mad2l1 | 0.267 | COX3 | -0.073 |
| cdk1 | 0.265 | b2ml | -0.072 |
| mki67 | 0.265 | rnasekb | -0.071 |
| dut | 0.264 | atp2b1a | -0.071 |
| fbxo5 | 0.262 | pvalb2 | -0.070 |
| cks1b | 0.259 | si:dkey-16p21.8 | -0.070 |
| aurkb | 0.258 | gapdhs | -0.069 |
| dek | 0.252 | nupr1a | -0.069 |
| top2a | 0.251 | ube2h | -0.066 |
| zgc:165555.3 | 0.247 | pvalb1 | -0.066 |
| rrm1 | 0.246 | XLOC-043857 | -0.066 |
| birc5a | 0.246 | nptna | -0.065 |
| si:dkey-261m9.17 | 0.240 | appb | -0.064 |
| plk1 | 0.240 | calm1b | -0.063 |
| cdca5 | 0.238 | cd164l2 | -0.063 |
| rrm2 | 0.237 | atp6ap2 | -0.063 |
| kifc1 | 0.237 | tmem59 | -0.063 |
| rpa2 | 0.232 | gpx2 | -0.062 |
| CABZ01058261.1 | 0.231 | apba1a | -0.062 |
| armc1l | 0.228 | atp1a3b | -0.062 |
| lbr | 0.227 | otofb | -0.061 |
| ube2c | 0.225 | gabarapl2 | -0.061 |
| hmgb2a | 0.224 | kif1aa | -0.061 |
| CABZ01005379.1 | 0.224 | ndrg3b | -0.060 |
| ccnb1 | 0.223 | CR925719.1 | -0.060 |
| sgo1 | 0.221 | ap3s1.1 | -0.060 |
| tubb2b | 0.221 | gb:bc139872 | -0.060 |
| cdc20 | 0.221 | aldocb | -0.060 |
| tuba8l | 0.220 | calm1a | -0.060 |
| cdca8 | 0.219 | tpi1a | -0.060 |
| si:ch211-69g19.2 | 0.218 | dedd1 | -0.060 |