Rac family small GTPase 1a
ZFIN 
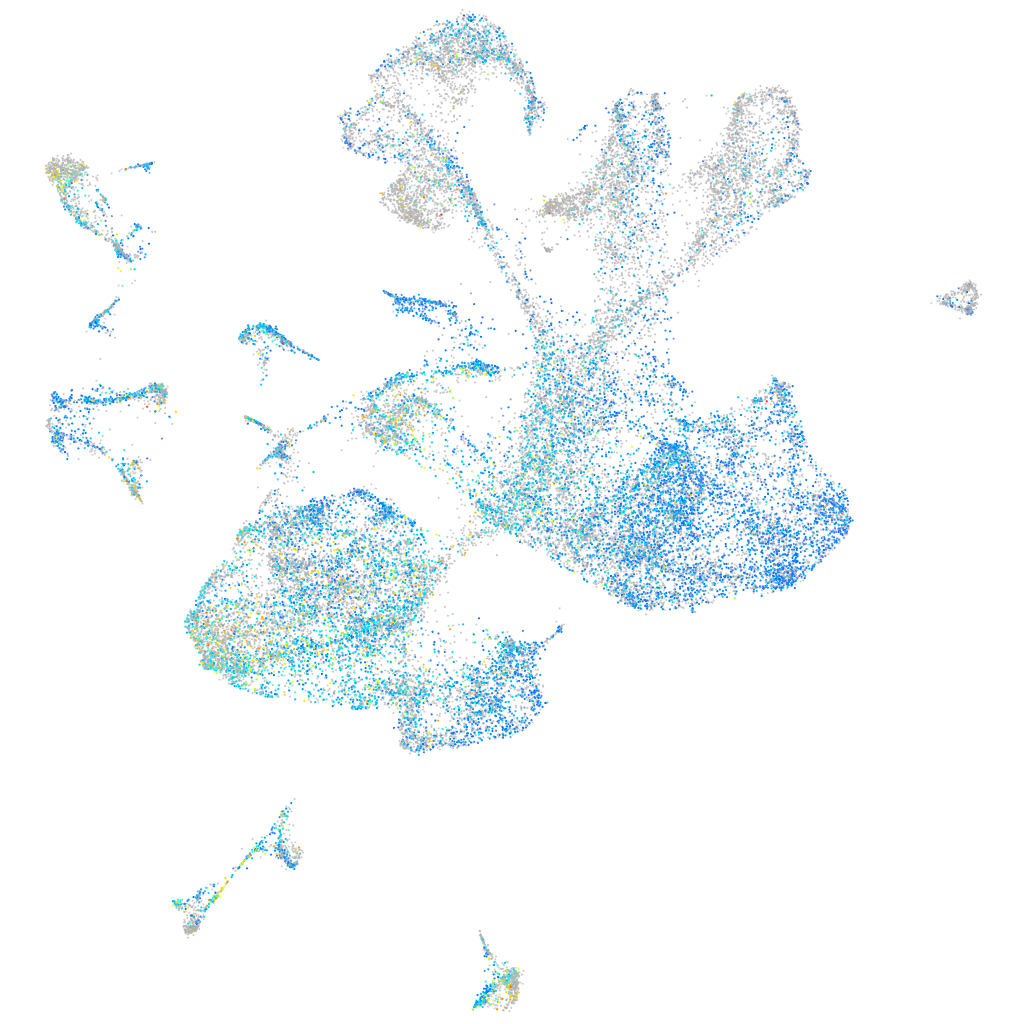
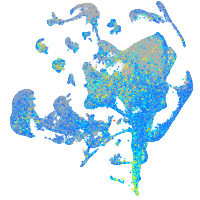
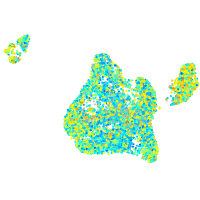
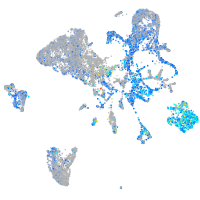
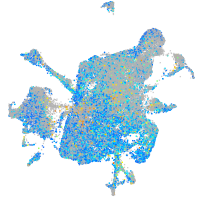
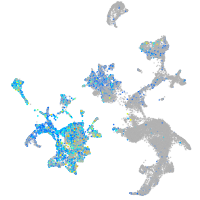
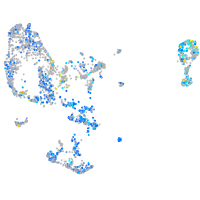
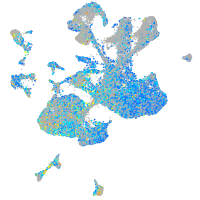
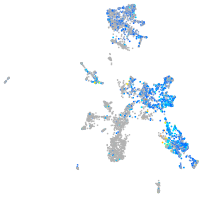
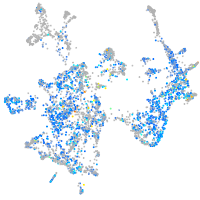
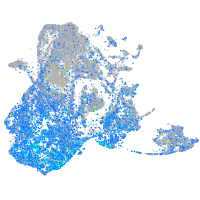
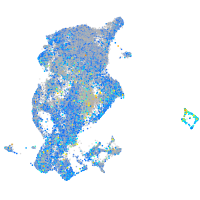
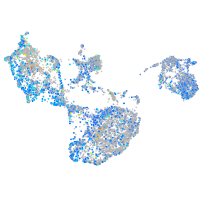
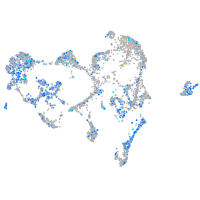
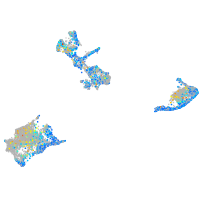
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.208 | elavl3 | -0.174 |
| cd82a | 0.187 | stxbp1a | -0.163 |
| fabp7a | 0.185 | elavl4 | -0.162 |
| actb2 | 0.183 | snap25a | -0.162 |
| msna | 0.180 | stx1b | -0.162 |
| psat1 | 0.178 | myt1b | -0.161 |
| sox2 | 0.178 | sncb | -0.159 |
| actb1 | 0.176 | vamp2 | -0.152 |
| msi1 | 0.174 | gng3 | -0.150 |
| notch3 | 0.174 | stmn1b | -0.150 |
| vamp3 | 0.171 | syt2a | -0.145 |
| gng5 | 0.168 | cplx2 | -0.145 |
| qki2 | 0.167 | tmem59l | -0.142 |
| gpm6bb | 0.165 | rbfox1 | -0.139 |
| cnn2 | 0.164 | sncgb | -0.138 |
| si:ch211-286b5.5 | 0.159 | nsg2 | -0.137 |
| gnai2a | 0.158 | atp6v0cb | -0.137 |
| XLOC-003690 | 0.157 | rtn1b | -0.131 |
| atp1b4 | 0.157 | cplx2l | -0.130 |
| tubb4b | 0.155 | rbfox3a | -0.130 |
| GCA | 0.155 | eno2 | -0.128 |
| klf6a | 0.153 | ptmaa | -0.128 |
| CR848047.1 | 0.151 | gng2 | -0.128 |
| si:ch1073-303k11.2 | 0.150 | pik3r3b | -0.127 |
| ccng1 | 0.148 | ywhag2 | -0.127 |
| ptprfb | 0.147 | jagn1a | -0.126 |
| cldn5a | 0.147 | stmn2a | -0.125 |
| canx | 0.147 | csdc2a | -0.124 |
| pgrmc1 | 0.146 | scrt2 | -0.124 |
| metrnla | 0.146 | LOC100537384 | -0.123 |
| CU467822.1 | 0.145 | ndrg4 | -0.122 |
| sox3 | 0.145 | onecut1 | -0.122 |
| sept10 | 0.142 | zc4h2 | -0.121 |
| psph | 0.142 | id4 | -0.121 |
| cd63 | 0.142 | aplp1 | -0.121 |