"RAB3C, member RAS oncogene family"
ZFIN 
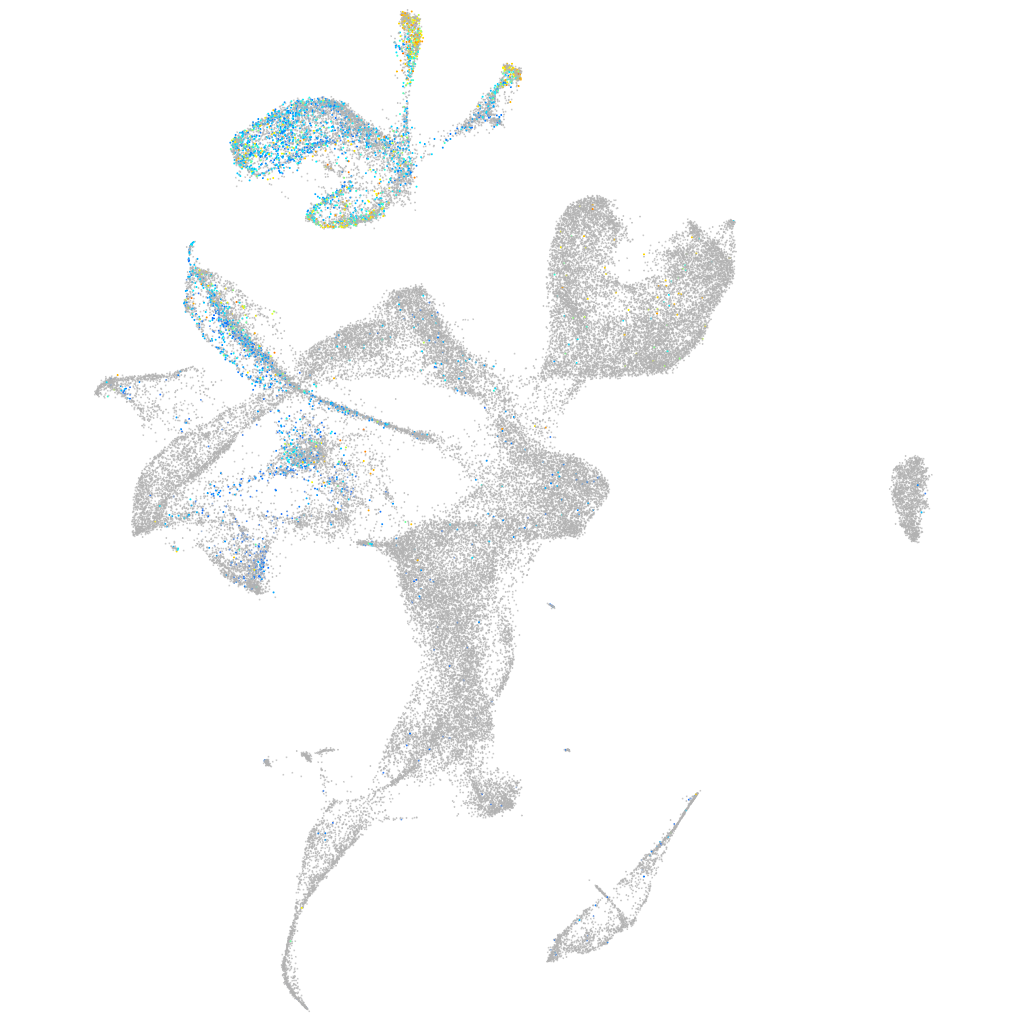
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| syt1a | 0.342 | hmgb2a | -0.231 |
| stxbp1a | 0.329 | pcna | -0.129 |
| slc32a1 | 0.313 | msna | -0.123 |
| ywhag2 | 0.305 | msi1 | -0.119 |
| stx1b | 0.293 | stmn1a | -0.119 |
| stmn1b | 0.279 | rrm1 | -0.118 |
| elavl3 | 0.276 | dut | -0.117 |
| mir181b-3 | 0.270 | hmga1a | -0.117 |
| tfap2a | 0.266 | nutf2l | -0.117 |
| rbfox1 | 0.266 | mki67 | -0.116 |
| gng3 | 0.265 | tuba8l | -0.115 |
| si:ch73-119p20.1 | 0.264 | mcm7 | -0.114 |
| nrxn1a | 0.264 | ahcy | -0.114 |
| sncb | 0.263 | ccna2 | -0.112 |
| slc6a1a | 0.252 | lbr | -0.112 |
| sv2a | 0.247 | chaf1a | -0.112 |
| barhl2 | 0.245 | ccnd1 | -0.111 |
| rtn1b | 0.242 | tuba8l4 | -0.110 |
| pvalb6 | 0.242 | her15.1 | -0.109 |
| tiam1a | 0.236 | dek | -0.106 |
| gabrb4 | 0.236 | mdka | -0.105 |
| eno2 | 0.233 | neurod4 | -0.105 |
| carmil2 | 0.232 | selenoh | -0.105 |
| st8sia5 | 0.231 | banf1 | -0.104 |
| zgc:153426 | 0.230 | crx | -0.103 |
| slc6a1b | 0.230 | eef1da | -0.103 |
| zgc:101840 | 0.228 | id1 | -0.103 |
| gnb1a | 0.228 | lig1 | -0.101 |
| pax6b | 0.226 | cks1b | -0.101 |
| gnao1a | 0.221 | otx5 | -0.101 |
| gldn | 0.220 | syt5b | -0.101 |
| zgc:65894 | 0.219 | rpa3 | -0.099 |
| dtnbp1b | 0.218 | rrm2 | -0.098 |
| rbfox2 | 0.218 | fen1 | -0.098 |
| palm1b | 0.217 | mad2l1 | -0.098 |