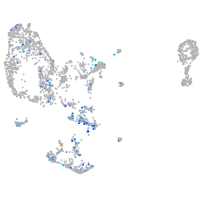
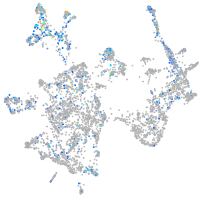
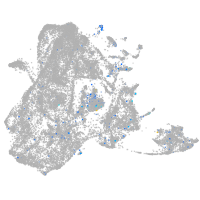
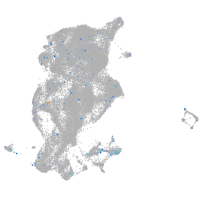
"RAB3A, member RAS oncogene family, a"
ZFIN 
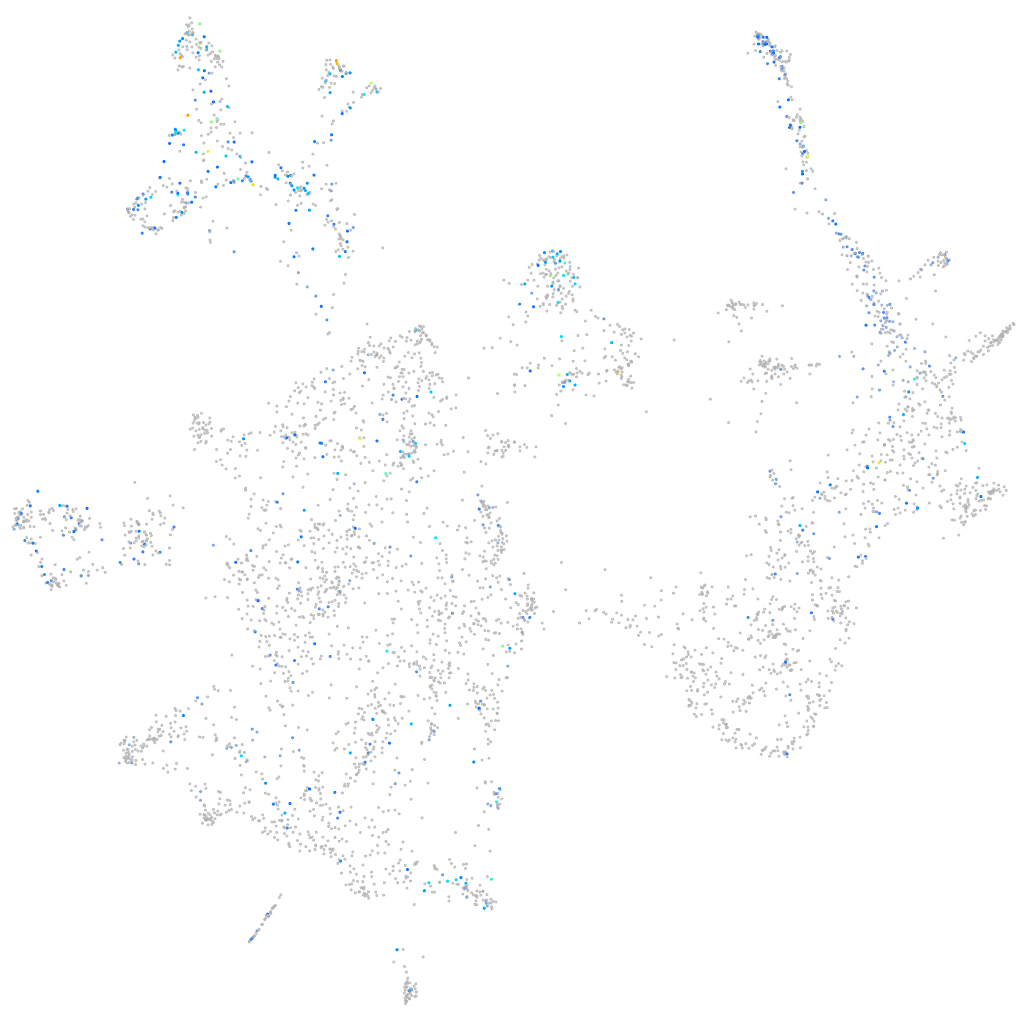
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrrc73 | 0.268 | rplp1 | -0.213 |
| atp1a3b | 0.260 | si:dkey-151g10.6 | -0.212 |
| nptna | 0.260 | rpl9 | -0.205 |
| pou4f1 | 0.259 | rplp2l | -0.200 |
| pcsk5a | 0.259 | tmsb4x | -0.197 |
| eef1a1b | 0.258 | rps25 | -0.192 |
| cd164l2 | 0.257 | rps20 | -0.191 |
| atp2b1a | 0.257 | rpl37 | -0.189 |
| dnajc5b | 0.257 | rps23 | -0.185 |
| tuba1a | 0.257 | rpl35a | -0.185 |
| osbpl1a | 0.256 | rps19 | -0.184 |
| tspan13b | 0.255 | rpl24 | -0.181 |
| emb | 0.255 | rps24 | -0.179 |
| pou4f3 | 0.254 | rps14 | -0.179 |
| gpx2 | 0.254 | rpl39 | -0.178 |
| gipc3 | 0.250 | rpl36a | -0.178 |
| otofb | 0.249 | rpl12 | -0.177 |
| myo6b | 0.249 | rps21 | -0.175 |
| gapdhs | 0.248 | rps2 | -0.174 |
| eps8b | 0.248 | rpl38 | -0.174 |
| spa17 | 0.247 | rpl23 | -0.170 |
| eno1a | 0.246 | rps10 | -0.170 |
| ccdc181 | 0.246 | rps28 | -0.168 |
| si:ch73-15n15.3 | 0.242 | rpl28 | -0.164 |
| si:ch211-284k5.2 | 0.242 | rpl35 | -0.164 |
| zgc:162989 | 0.242 | rpl26 | -0.163 |
| atp1b2b | 0.241 | rps27.1 | -0.162 |
| calml4a | 0.241 | rps8a | -0.161 |
| syt14b | 0.240 | rps11 | -0.161 |
| ckbb | 0.239 | rpl29 | -0.160 |
| slc17a8 | 0.239 | rpl34 | -0.160 |
| CR925719.1 | 0.238 | faua | -0.158 |
| rnasekb | 0.238 | rps12 | -0.157 |
| grxcr2 | 0.237 | epcam | -0.156 |
| dnah9l | 0.237 | rpl31 | -0.155 |