"RAB3A, member RAS oncogene family, a"
ZFIN 
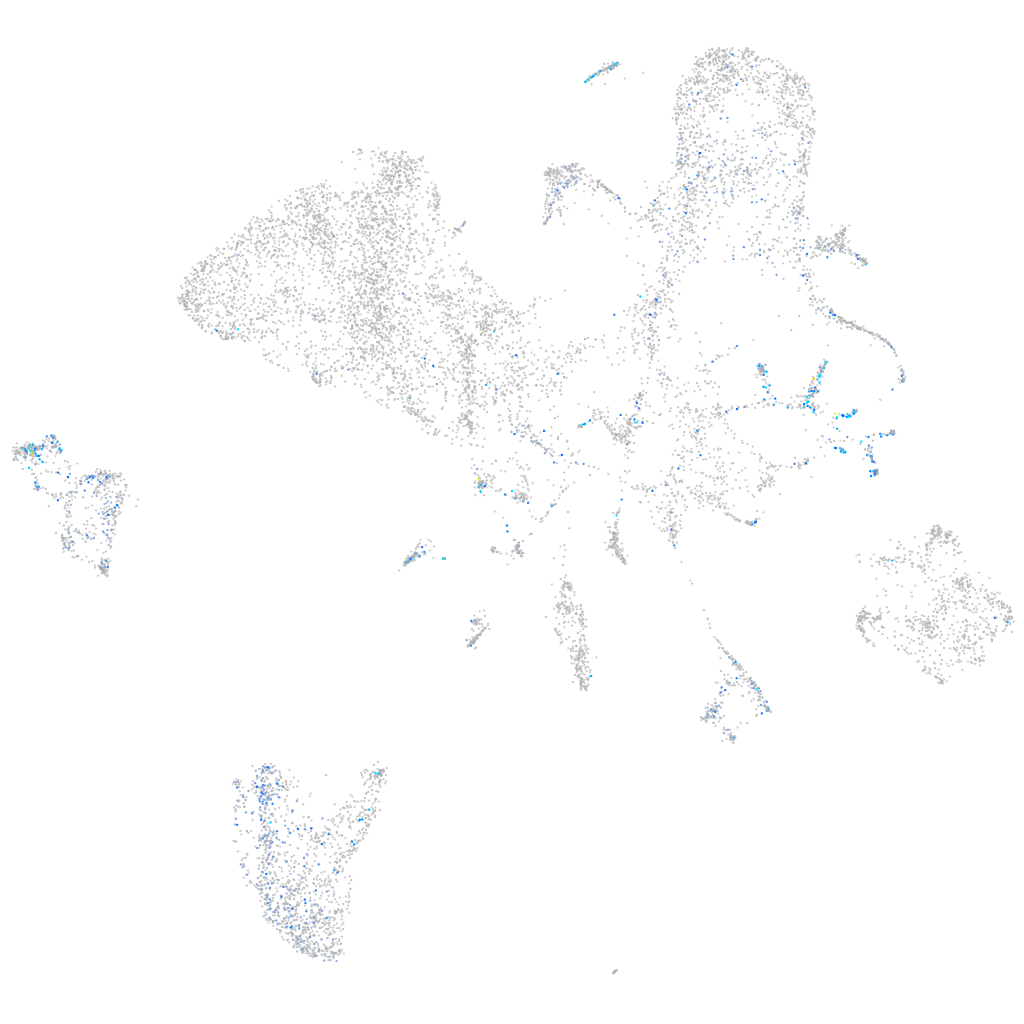
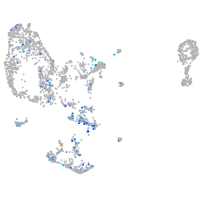
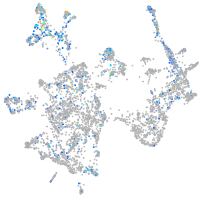
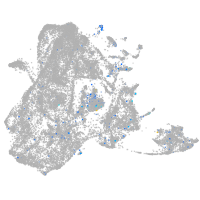
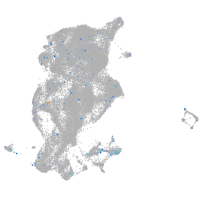
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.288 | apoc1 | -0.174 |
| pcsk1nl | 0.267 | apoa4b.1 | -0.163 |
| scgn | 0.262 | apoa1b | -0.162 |
| atp6v0cb | 0.262 | afp4 | -0.159 |
| calm1b | 0.259 | gapdh | -0.154 |
| neurod1 | 0.258 | apoc2 | -0.152 |
| scg2a | 0.245 | apoa2 | -0.150 |
| gapdhs | 0.245 | fetub | -0.145 |
| fev | 0.244 | rbp2b | -0.145 |
| mir375-2 | 0.244 | serpina1 | -0.145 |
| rnasekb | 0.243 | serpina1l | -0.145 |
| pax6b | 0.239 | fabp10a | -0.145 |
| hpcal1 | 0.238 | apom | -0.145 |
| rasd4 | 0.236 | tfa | -0.144 |
| si:dkey-153k10.9 | 0.235 | ambp | -0.144 |
| zgc:101731 | 0.235 | ces2 | -0.144 |
| vat1 | 0.234 | zgc:123103 | -0.143 |
| mir7a-1 | 0.233 | si:dkey-16p21.8 | -0.142 |
| rprmb | 0.233 | gamt | -0.142 |
| syt1a | 0.233 | rbp4 | -0.142 |
| insm1a | 0.227 | fgg | -0.142 |
| insm1b | 0.224 | mgst1.2 | -0.142 |
| tspan7b | 0.220 | scp2a | -0.141 |
| nucb2b | 0.219 | hao1 | -0.141 |
| hepacam2 | 0.218 | gc | -0.140 |
| pcsk2 | 0.216 | ttr | -0.140 |
| tmed4 | 0.216 | LOC110437731 | -0.140 |
| kcnj11 | 0.210 | apobb.1 | -0.139 |
| calm1a | 0.210 | ftcd | -0.138 |
| atpv0e2 | 0.209 | tdo2a | -0.138 |
| isl1 | 0.207 | fgb | -0.138 |
| slc1a4 | 0.206 | uox | -0.137 |
| id4 | 0.206 | hpda | -0.137 |
| vamp2 | 0.204 | zgc:112265 | -0.137 |
| tmed7 | 0.203 | fbp1b | -0.136 |