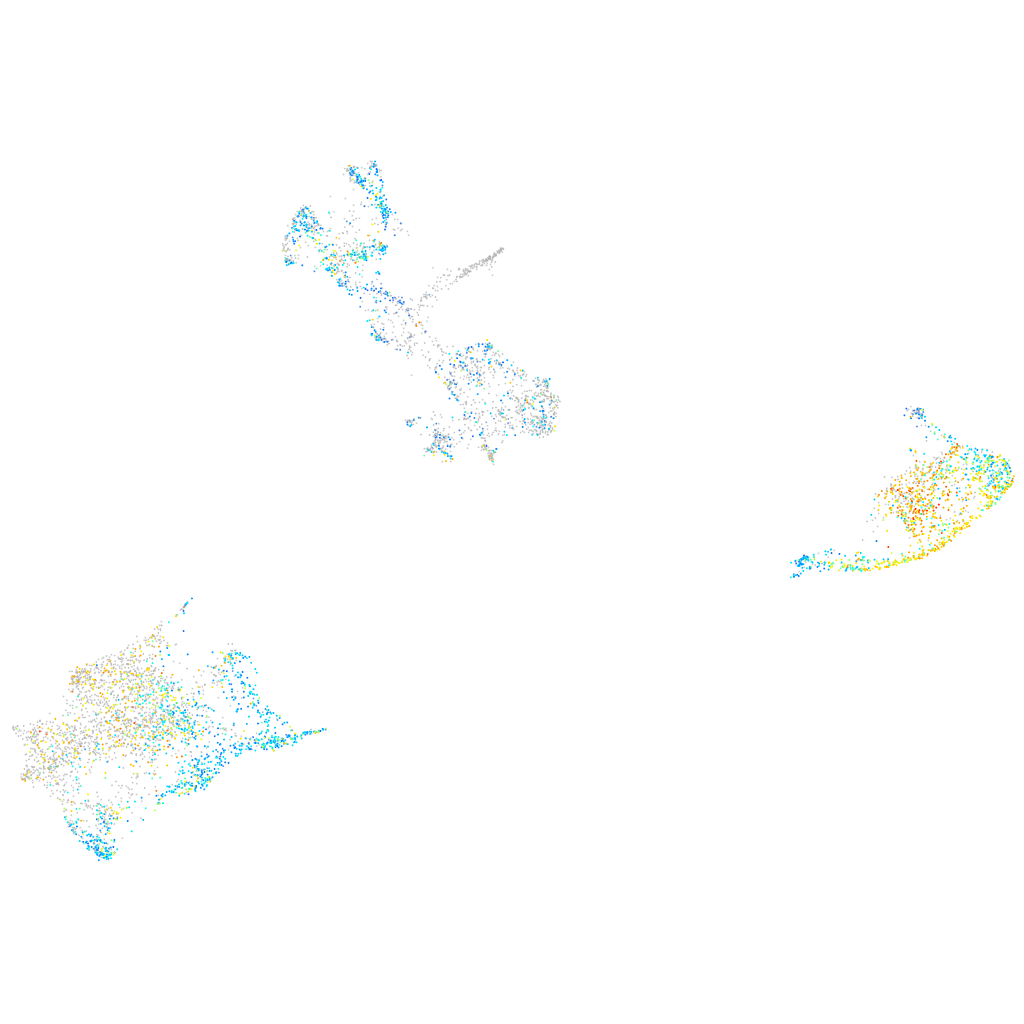
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.465 | ptmaa | -0.220 |
| pmela | 0.462 | si:ch211-222l21.1 | -0.186 |
| tspan36 | 0.458 | hmgb1b | -0.182 |
| zgc:91968 | 0.457 | si:ch73-1a9.3 | -0.180 |
| tyrp1b | 0.455 | CABZ01021592.1 | -0.179 |
| tyrp1a | 0.451 | marcksl1a | -0.178 |
| tyr | 0.440 | hmgn2 | -0.176 |
| slc45a2 | 0.435 | gpm6aa | -0.174 |
| oca2 | 0.414 | nova2 | -0.168 |
| qdpra | 0.408 | tuba1c | -0.167 |
| slc24a5 | 0.405 | stmn1b | -0.163 |
| mtbl | 0.393 | hmgb3a | -0.156 |
| slc37a2 | 0.390 | mdkb | -0.155 |
| tspan10 | 0.388 | stmn1a | -0.152 |
| si:ch73-389b16.1 | 0.388 | pvalb2 | -0.147 |
| pah | 0.378 | hmgb2b | -0.146 |
| rabl6b | 0.372 | ptmab | -0.146 |
| atp6v0ca | 0.371 | dlx5a | -0.144 |
| gstt1a | 0.369 | elavl3 | -0.141 |
| slc3a2a | 0.360 | gng3 | -0.140 |
| kita | 0.354 | epb41a | -0.138 |
| prkar1b | 0.353 | sncb | -0.136 |
| mitfa | 0.352 | gng2 | -0.136 |
| tfap2e | 0.350 | h3f3a | -0.136 |
| tmem243b | 0.349 | hmgb2a | -0.136 |
| anxa1a | 0.337 | si:ch73-281n10.2 | -0.135 |
| lamp1a | 0.335 | zc4h2 | -0.135 |
| aadac | 0.330 | tmsb | -0.134 |
| gpr143 | 0.330 | acbd7 | -0.131 |
| bace2 | 0.326 | pvalb1 | -0.131 |
| syngr1a | 0.322 | mdka | -0.129 |
| mlpha | 0.319 | FO082781.1 | -0.128 |
| rab32a | 0.312 | CU467822.1 | -0.127 |
| atp6v0a2b | 0.309 | uraha | -0.126 |
| tmem243a | 0.309 | cx43.4 | -0.126 |