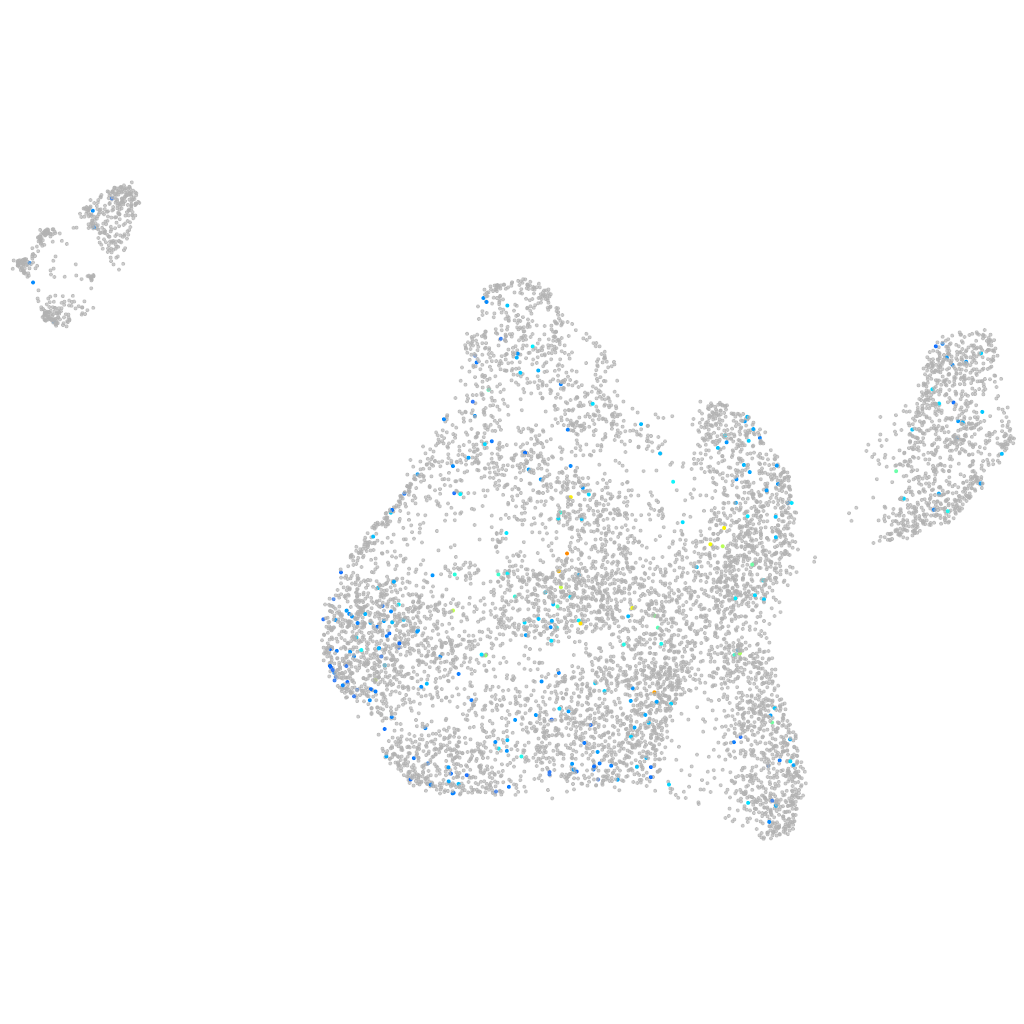
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm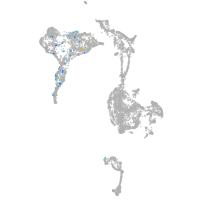 |
muscle 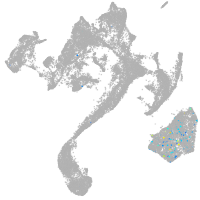 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 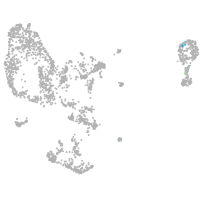 |
neural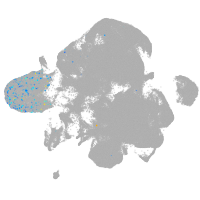 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcna7 | 0.102 | h1m | -0.039 |
| FP085399.4 | 0.099 | h2af1al | -0.038 |
| CCR8 | 0.097 | smad2 | -0.036 |
| myl4 | 0.095 | zgc:162025 | -0.035 |
| a1cf | 0.091 | arhgdig | -0.035 |
| LOC797394 | 0.089 | ell | -0.034 |
| LOC110439602 | 0.088 | casp8ap2 | -0.034 |
| galnt14 | 0.088 | ciarta | -0.034 |
| LOC110439797 | 0.086 | inip | -0.033 |
| LOC570163 | 0.086 | clns1a | -0.033 |
| si:dkey-237j11.3 | 0.086 | si:dkeyp-2e4.2 | -0.032 |
| tspearb | 0.084 | cyp11a1 | -0.032 |
| rhpn1 | 0.083 | cnot2 | -0.032 |
| krt1-19d | 0.083 | NC-002333.4 | -0.032 |
| cacnb4b | 0.081 | si:ch211-210c8.7 | -0.032 |
| agtr1b | 0.079 | eloal | -0.031 |
| BX571942.1 | 0.077 | ezrb | -0.031 |
| CT030005.2 | 0.076 | snx10a | -0.031 |
| CR626886.2 | 0.076 | tob1a | -0.031 |
| vav3 | 0.075 | pafah1b1a | -0.031 |
| cyp11a2 | 0.074 | nupr1a | -0.031 |
| fam168a | 0.074 | lipg | -0.031 |
| sparcl2 | 0.073 | sec24c | -0.031 |
| kcnb1 | 0.073 | acp5a | -0.031 |
| CR931780.1 | 0.073 | RNF219 | -0.031 |
| dao.3 | 0.073 | cnot3b | -0.030 |
| slc7a7 | 0.072 | gosr1 | -0.030 |
| pdzrn3a | 0.072 | brd2b | -0.030 |
| arhgap29a | 0.071 | exosc7 | -0.030 |
| lrrc74a | 0.071 | mknk2a | -0.030 |
| LOC101883552 | 0.071 | cdca7b | -0.030 |
| si:dkey-220o5.5 | 0.071 | zfand5b | -0.030 |
| BX088587.1 | 0.070 | hebp2 | -0.030 |
| smoc2 | 0.070 | mmaa | -0.029 |
| cd81b | 0.070 | dpf2 | -0.029 |