glutamine-rich 1
ZFIN 
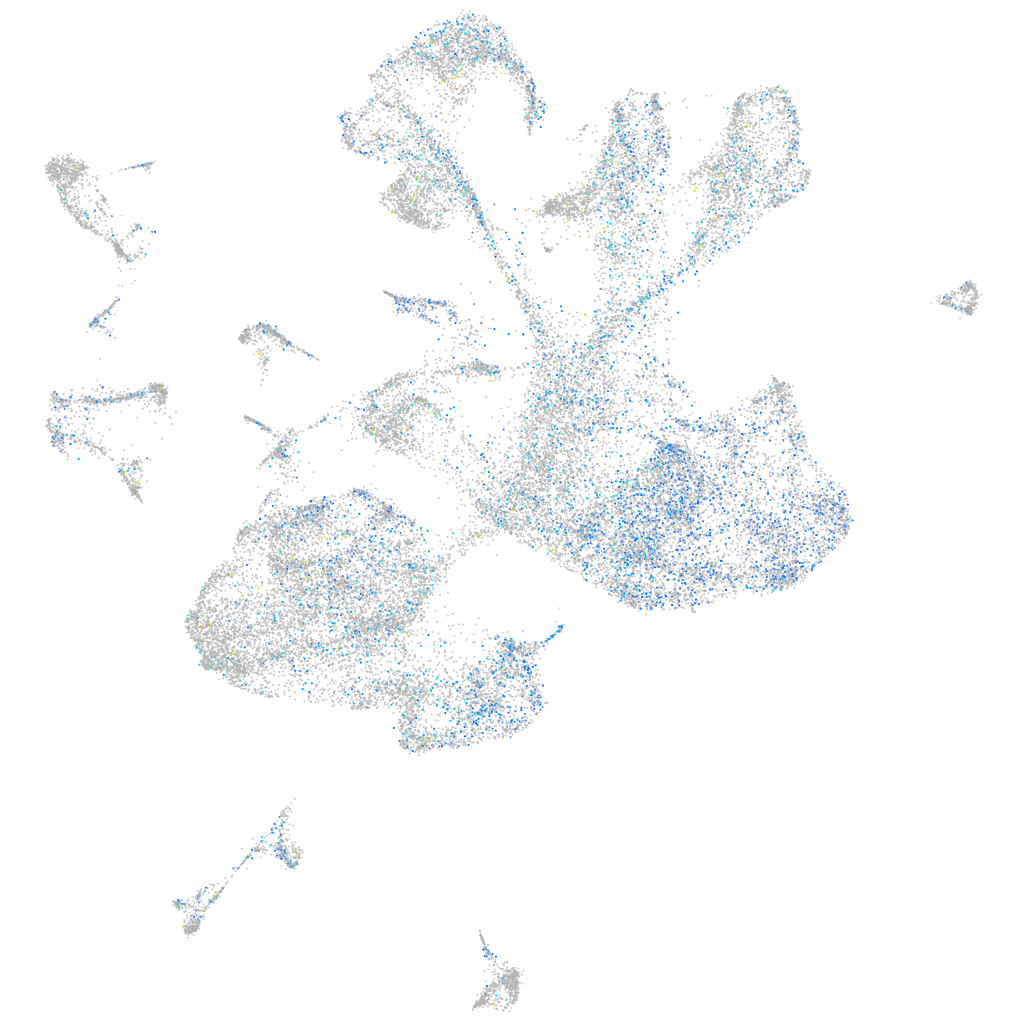
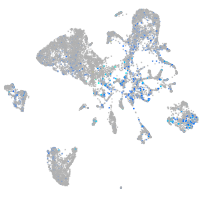
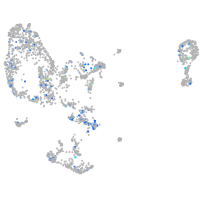
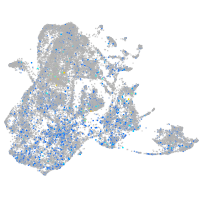
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tubb2b | 0.102 | slc1a2b | -0.057 |
| h3f3a | 0.098 | cx43 | -0.055 |
| h2afvb | 0.095 | ptn | -0.054 |
| hmga1a | 0.095 | glula | -0.053 |
| seta | 0.093 | atp1a1b | -0.053 |
| cirbpa | 0.092 | efhd1 | -0.052 |
| hmgb2b | 0.092 | si:ch211-66e2.5 | -0.051 |
| hnrnpaba | 0.091 | slc4a4a | -0.051 |
| ran | 0.091 | mt2 | -0.050 |
| khdrbs1a | 0.091 | acbd7 | -0.049 |
| hnrnpabb | 0.090 | mdkb | -0.049 |
| snrpf | 0.089 | fabp7a | -0.048 |
| snrpd1 | 0.088 | pvalb1 | -0.048 |
| cbx3a | 0.088 | gpr37l1b | -0.048 |
| snrpe | 0.088 | CR383676.1 | -0.046 |
| hdac1 | 0.087 | pvalb2 | -0.045 |
| hmgb2a | 0.087 | hepacama | -0.045 |
| si:ch211-222l21.1 | 0.086 | ppap2d | -0.045 |
| ddx39ab | 0.085 | cox4i2 | -0.045 |
| snrpd2 | 0.085 | slc3a2a | -0.045 |
| hnrnpa0l | 0.085 | gapdhs | -0.045 |
| dut | 0.084 | apoa1b | -0.045 |
| rrm1 | 0.084 | ckbb | -0.045 |
| tuba8l4 | 0.084 | aldocb | -0.044 |
| si:ch211-288g17.3 | 0.083 | ndrg3a | -0.043 |
| hnrnpa0b | 0.082 | slc1a3b | -0.043 |
| pcna | 0.082 | ak1 | -0.043 |
| rrm2 | 0.082 | lpin1 | -0.042 |
| nono | 0.081 | qki2 | -0.041 |
| alyref | 0.081 | slc6a1b | -0.041 |
| ptmab | 0.080 | s100b | -0.040 |
| hmgn2 | 0.080 | rpz5 | -0.040 |
| snrpb | 0.080 | cd63 | -0.040 |
| setb | 0.080 | slc6a11b | -0.040 |
| rbbp4 | 0.079 | anxa13 | -0.040 |