glutaminyl-peptide cyclotransferase-like b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 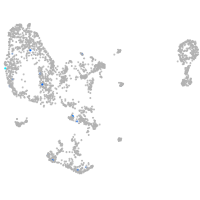 |
neural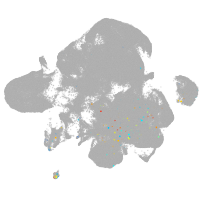 |
spinal cord / glia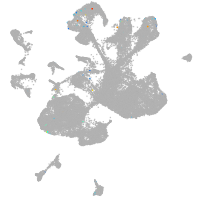 |
eye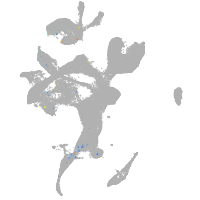 |
taste / olfactory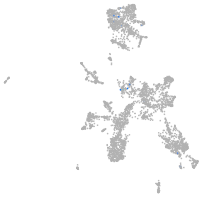 |
otic / lateral line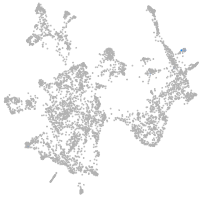 |
epidermis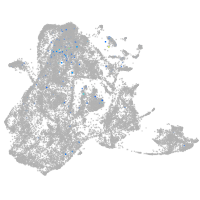 |
periderm |
fin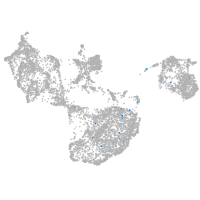 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-119p20.1 | 0.064 | hspb1 | -0.024 |
| si:ch211-256e16.6 | 0.061 | cx43.4 | -0.022 |
| ndufa4 | 0.059 | akap12b | -0.020 |
| eno2 | 0.058 | pou5f3 | -0.020 |
| sncgb | 0.058 | s100a1 | -0.020 |
| atp2b3b | 0.057 | stm | -0.020 |
| CABZ01073083.1 | 0.057 | si:ch211-152c2.3 | -0.019 |
| gfral | 0.057 | apoc1 | -0.018 |
| grp | 0.057 | asb11 | -0.018 |
| cplx2 | 0.054 | ccnb1 | -0.018 |
| vsnl1b | 0.054 | hnrnpa1b | -0.018 |
| snap25a | 0.053 | mki67 | -0.018 |
| sv2a | 0.053 | msna | -0.018 |
| syn2a | 0.052 | si:dkey-66i24.9 | -0.018 |
| atp2b3a | 0.051 | tpx2 | -0.018 |
| CR759836.1 | 0.051 | vox | -0.018 |
| gapdhs | 0.051 | zgc:56699 | -0.018 |
| ywhag2 | 0.051 | banf1 | -0.017 |
| calm1b | 0.050 | ccna2 | -0.017 |
| stx1b | 0.050 | dkc1 | -0.017 |
| stxbp1a | 0.050 | lig1 | -0.017 |
| nptna | 0.049 | nnr | -0.017 |
| syt2a | 0.049 | polr3gla | -0.017 |
| map1aa | 0.048 | si:ch1073-80i24.3 | -0.017 |
| sncb | 0.048 | tbx16 | -0.017 |
| zgc:112408 | 0.048 | ved | -0.017 |
| elavl4 | 0.047 | aldob | -0.016 |
| stmn2a | 0.047 | arf1 | -0.016 |
| syt1a | 0.047 | bms1 | -0.016 |
| atp1b1b | 0.045 | cd2bp2 | -0.016 |
| atp6v0cb | 0.045 | chaf1a | -0.016 |
| rab6bb | 0.045 | fbl | -0.016 |
| tmem42b | 0.045 | fbxo5 | -0.016 |
| vamp2 | 0.045 | gar1 | -0.016 |
| zgc:65894 | 0.045 | nop58 | -0.016 |




