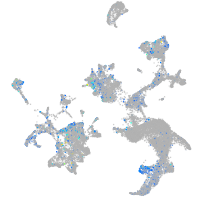
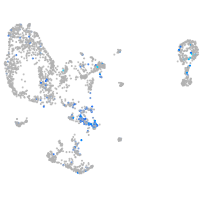
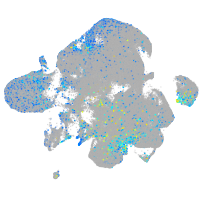
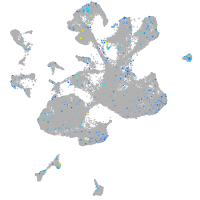
quinoid dihydropteridine reductase a
ZFIN 
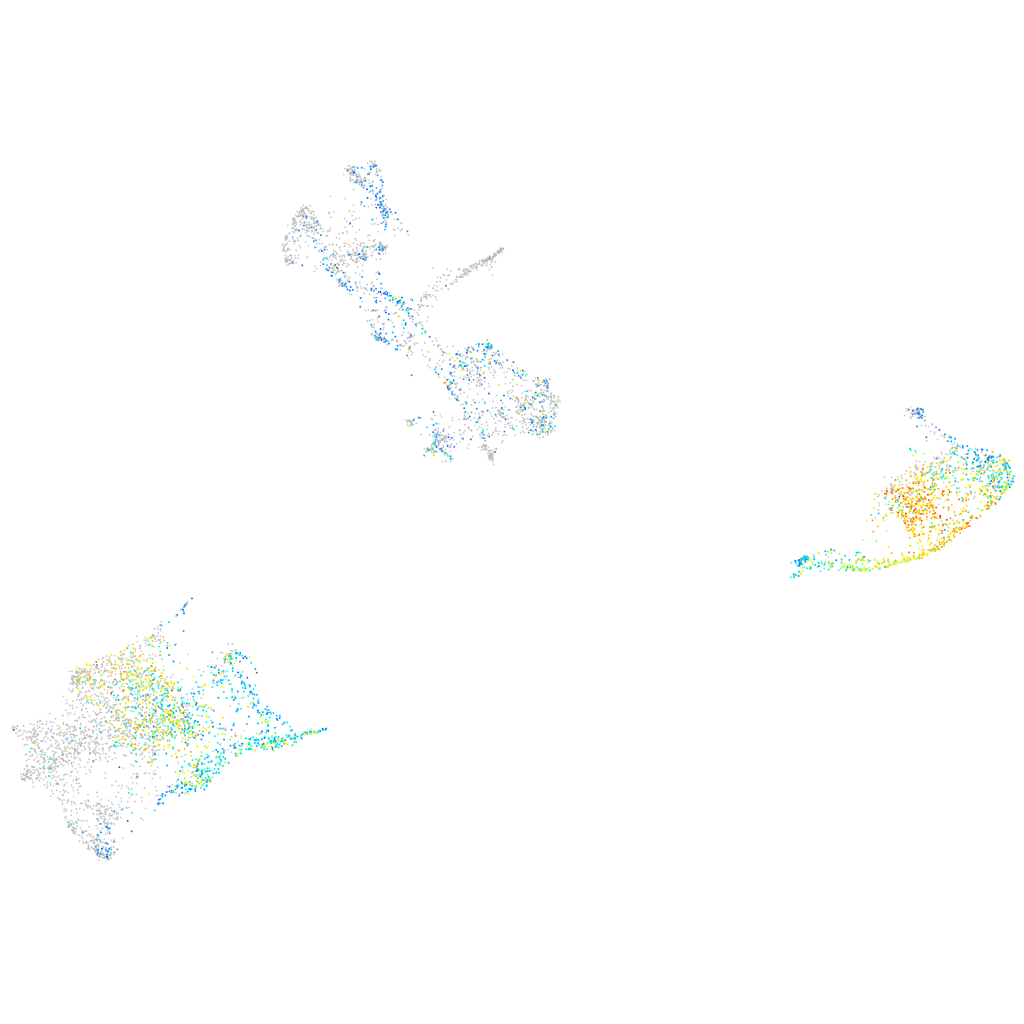
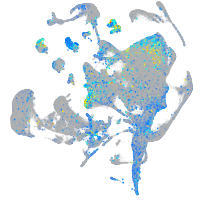
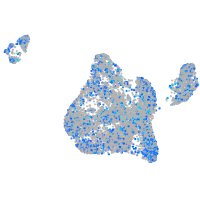
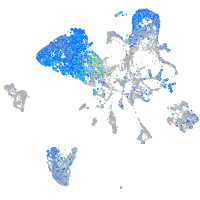
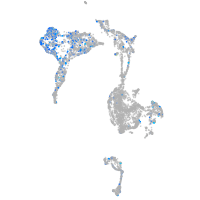
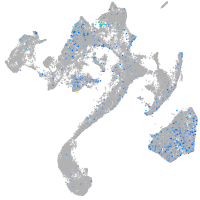
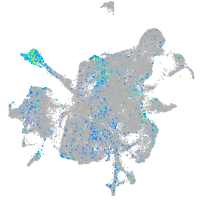
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pah | 0.511 | defbl1 | -0.285 |
| dct | 0.484 | apoda.1 | -0.278 |
| tyrp1a | 0.477 | gpnmb | -0.261 |
| pmela | 0.474 | si:ch211-256m1.8 | -0.259 |
| zgc:91968 | 0.471 | lypc | -0.255 |
| tyrp1b | 0.462 | mdkb | -0.253 |
| pcbd1 | 0.461 | si:dkey-197i20.6 | -0.235 |
| tyr | 0.447 | mdka | -0.233 |
| gstt1a | 0.438 | s100v2 | -0.224 |
| tspan36 | 0.438 | akap12a | -0.224 |
| slc37a2 | 0.426 | si:ch73-89b15.3 | -0.222 |
| oca2 | 0.415 | ppdpfb | -0.221 |
| tspan10 | 0.412 | apoa2 | -0.219 |
| rab38 | 0.408 | unm-sa821 | -0.218 |
| slc24a5 | 0.405 | alx4a | -0.214 |
| mitfa | 0.400 | sesn1 | -0.208 |
| rabl6b | 0.400 | ponzr1 | -0.207 |
| mtbl | 0.396 | slc25a36a | -0.204 |
| slc45a2 | 0.391 | alx4b | -0.200 |
| atp6v0ca | 0.390 | prx | -0.197 |
| tmem243b | 0.385 | sdc2 | -0.196 |
| si:ch73-389b16.1 | 0.378 | alx1 | -0.196 |
| tmem243a | 0.377 | sytl2b | -0.195 |
| prkar1b | 0.366 | guk1a | -0.193 |
| C12orf75 | 0.364 | dap1b | -0.193 |
| syngr1a | 0.360 | krt18b | -0.192 |
| abracl | 0.355 | agmo | -0.192 |
| anxa1a | 0.348 | tcirg1a | -0.191 |
| tfap2e | 0.346 | si:ch211-105c13.3 | -0.190 |
| aadac | 0.342 | ptmaa | -0.190 |
| atp6v1g1 | 0.336 | pvalb1 | -0.188 |
| lamp1a | 0.333 | ndrg2 | -0.188 |
| slc3a2a | 0.330 | cdh11 | -0.187 |
| slc29a3 | 0.329 | apoa1b | -0.185 |
| rab32a | 0.325 | pvalb2 | -0.184 |