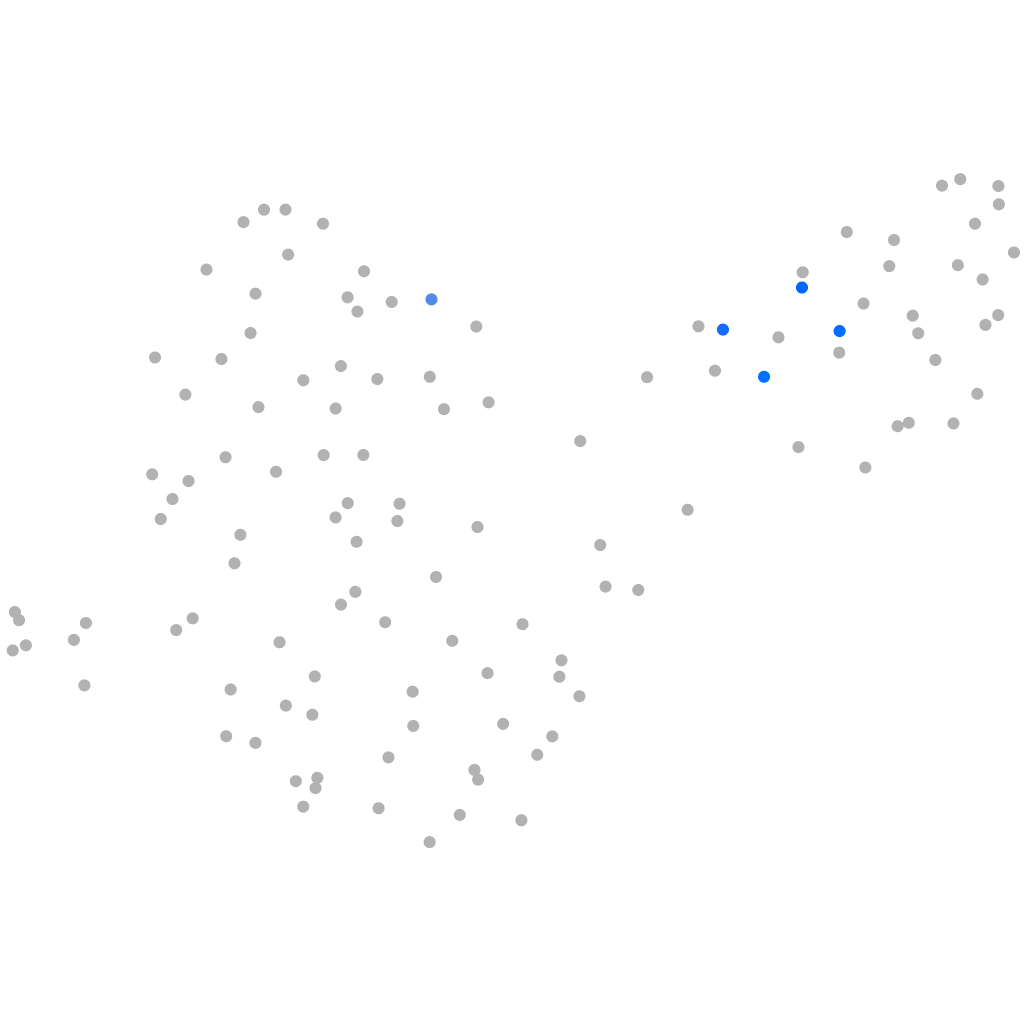
"pentraxin 4, long"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| radx | 0.798 | hnrnpaba | -0.271 |
| tfap2d | 0.714 | brd3a | -0.270 |
| tdp2a | 0.686 | snrpa | -0.261 |
| FRMD1 | 0.674 | hmgb1a | -0.238 |
| si:ch73-182a11.2 | 0.674 | akap12b | -0.227 |
| si:dkey-9c18.3 | 0.654 | nop56 | -0.222 |
| chst3a | 0.650 | ybx1 | -0.218 |
| tbx22 | 0.647 | ptmab | -0.211 |
| zgc:153039 | 0.640 | srrt | -0.205 |
| c2cd3 | 0.629 | elavl1a | -0.204 |
| st6galnac3 | 0.619 | chd4b | -0.203 |
| smc1a | 0.619 | tpx2 | -0.200 |
| hephl1a | 0.614 | apoeb | -0.200 |
| gcnt4a | 0.606 | ppig | -0.197 |
| slc24a3 | 0.583 | npm1a | -0.195 |
| si:dkey-262g12.14 | 0.578 | ncl | -0.194 |
| pcp4a | 0.574 | hnrnpa1a | -0.193 |
| sfxn5b | 0.570 | h1m | -0.190 |
| nek12 | 0.568 | kif23 | -0.189 |
| pkhd1l1 | 0.568 | ddx23 | -0.187 |
| itm2ca | 0.565 | si:ch73-281n10.2 | -0.186 |
| hbbe3 | 0.564 | metap2b | -0.185 |
| slit1b | 0.561 | max | -0.184 |
| gucy2d | 0.559 | nucks1a | -0.184 |
| dbx1b | 0.557 | tbx16 | -0.183 |
| crip3 | 0.551 | nusap1 | -0.181 |
| tgfb2 | 0.548 | hnrnpa1b | -0.181 |
| FP017215.1 | 0.547 | NC-002333.4 | -0.180 |
| LOC110439111 | 0.547 | tcea1 | -0.180 |
| mef2ca | 0.544 | epcam | -0.179 |
| bsnb | 0.543 | si:ch211-51e12.7 | -0.178 |
| faxcb | 0.543 | ccna2 | -0.177 |
| lrp1ba | 0.543 | tmed2 | -0.177 |
| nptx2b | 0.543 | hspd1 | -0.176 |
| sema7a | 0.543 | marcksb | -0.175 |