prothymosin alpha a
ZFIN 
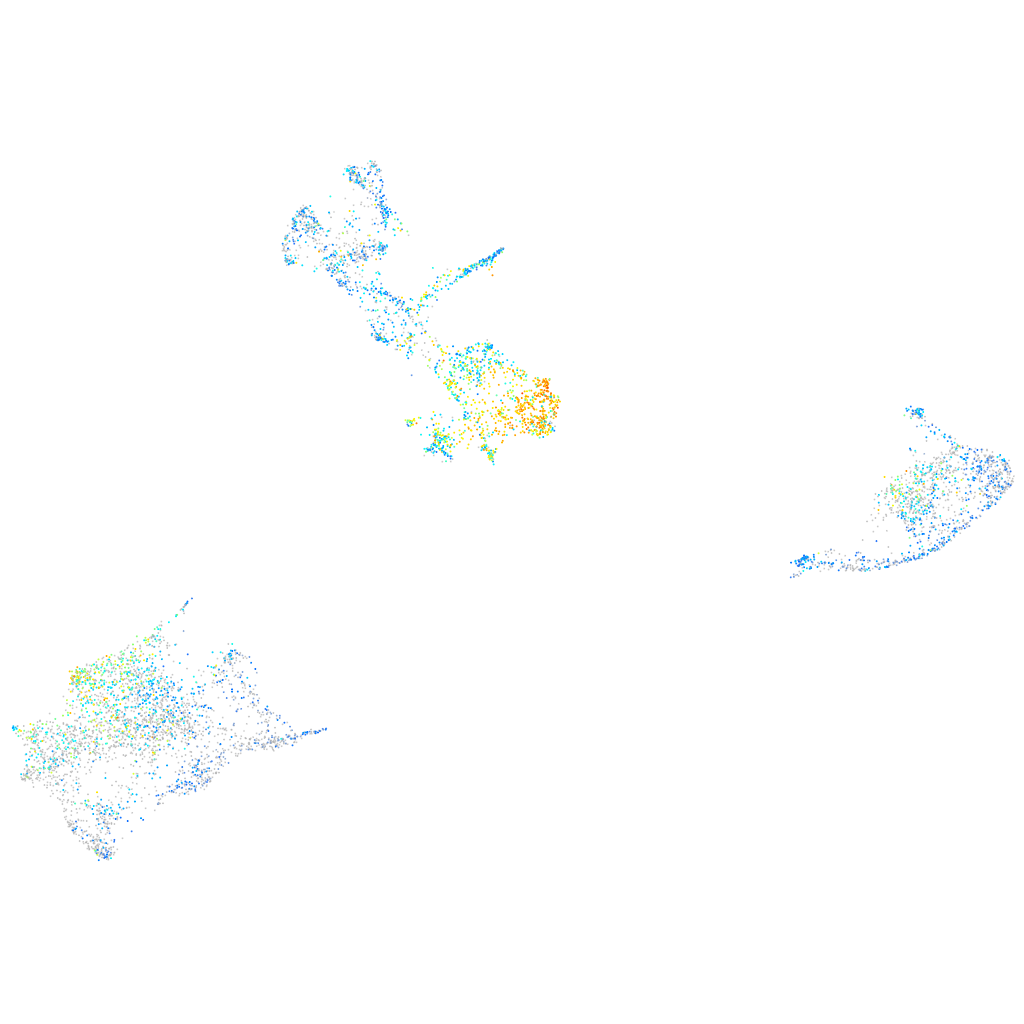
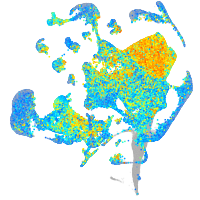
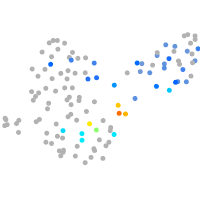
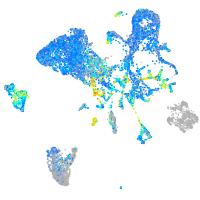
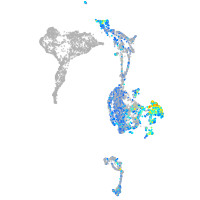
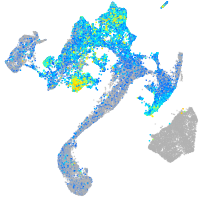
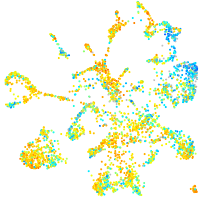
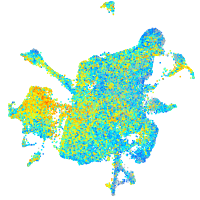
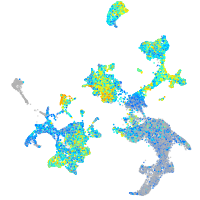
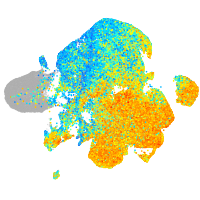
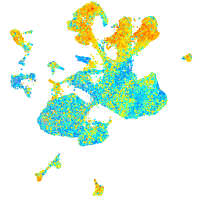
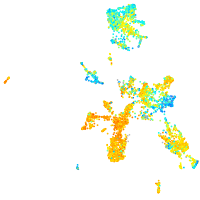
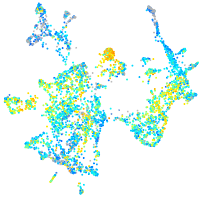
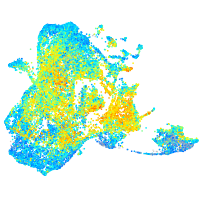
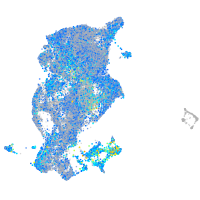
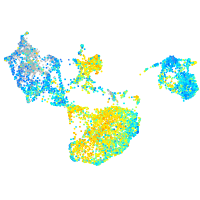
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.507 | tspan36 | -0.280 |
| tuba1c | 0.498 | syngr1a | -0.266 |
| nova2 | 0.489 | gstp1 | -0.259 |
| hmgb1b | 0.481 | akr1b1 | -0.243 |
| stmn1b | 0.476 | pts | -0.238 |
| ptmab | 0.476 | LOC103910009 | -0.234 |
| si:ch211-222l21.1 | 0.476 | gpr143 | -0.229 |
| elavl3 | 0.472 | actb2 | -0.225 |
| si:ch73-1a9.3 | 0.437 | gch2 | -0.224 |
| zc4h2 | 0.430 | pttg1ipb | -0.220 |
| hmgb3a | 0.427 | rab38 | -0.220 |
| tmsb | 0.415 | slc22a7a | -0.218 |
| gng3 | 0.407 | pcbd1 | -0.217 |
| rnasekb | 0.405 | impdh1b | -0.212 |
| marcksl1a | 0.405 | rnaseka | -0.208 |
| epb41a | 0.399 | C12orf75 | -0.208 |
| tmeff1b | 0.395 | glulb | -0.203 |
| sncb | 0.390 | mitfa | -0.201 |
| chd4a | 0.388 | cx30.3 | -0.200 |
| rtn1a | 0.384 | bace2 | -0.200 |
| marcksb | 0.380 | tmem130 | -0.196 |
| hmgn2 | 0.375 | tdh | -0.196 |
| tubb5 | 0.369 | trpm1b | -0.195 |
| h3f3a | 0.368 | si:dkey-21a6.5 | -0.194 |
| si:ch211-288g17.3 | 0.368 | gpd1b | -0.194 |
| tubb2b | 0.366 | rabl6b | -0.192 |
| fez1 | 0.365 | qdpra | -0.190 |
| cadm3 | 0.358 | slc2a11b | -0.190 |
| hnrnpaba | 0.356 | paics | -0.190 |
| vamp2 | 0.356 | rab32a | -0.189 |
| elavl4 | 0.356 | slc2a15a | -0.188 |
| CU467822.1 | 0.348 | zgc:110239 | -0.188 |
| tuba1a | 0.346 | rab34b | -0.187 |
| gng2 | 0.344 | sprb | -0.187 |
| tox | 0.342 | ahnak | -0.186 |