prostaglandin-endoperoxide synthase 1
ZFIN 
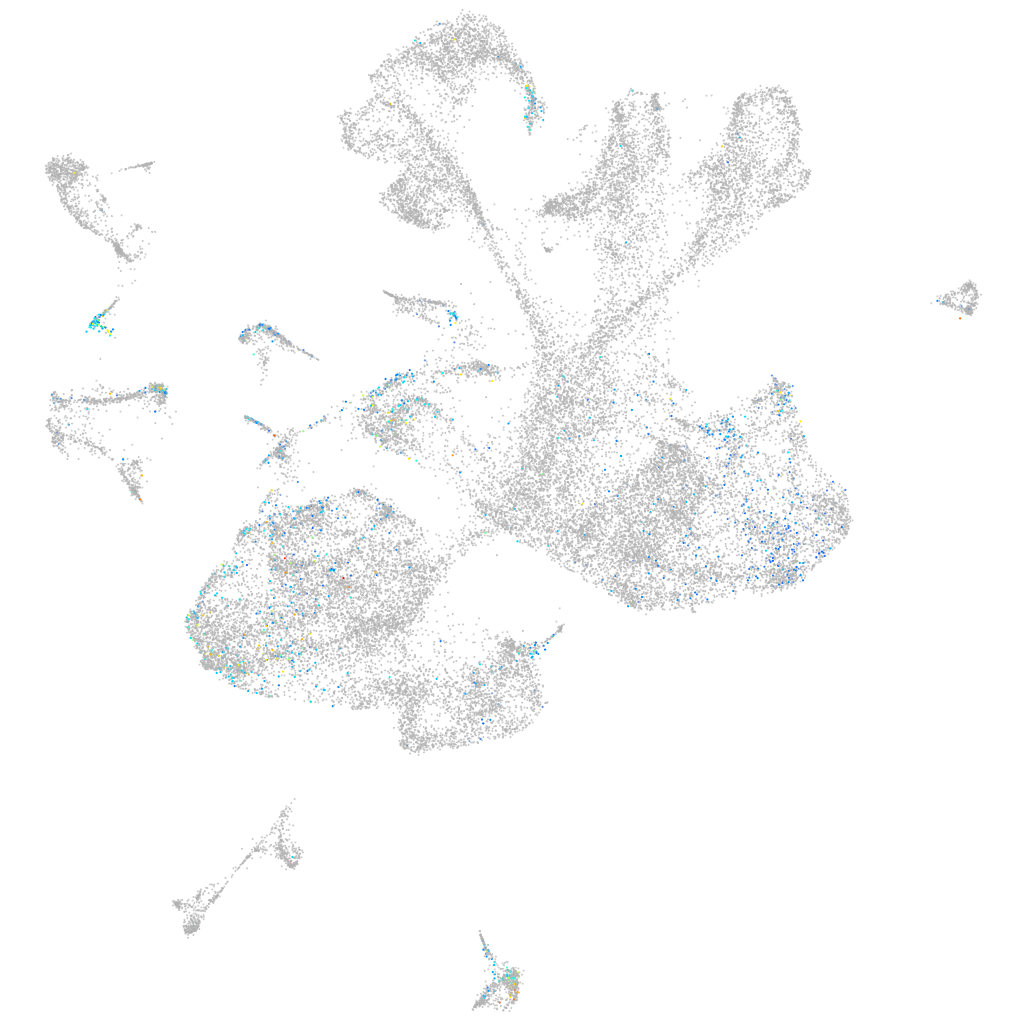
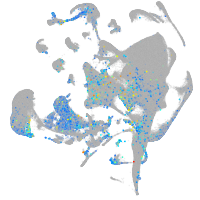
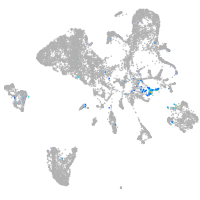
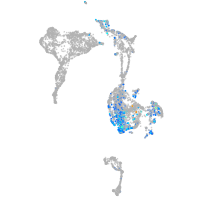
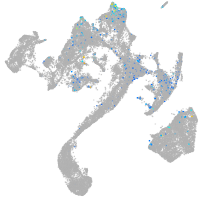
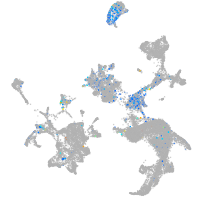
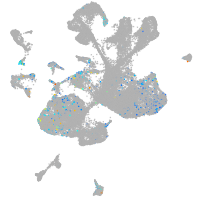
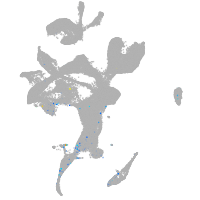
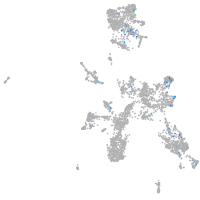
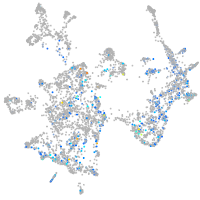
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-157b11.8 | 0.156 | fabp3 | -0.092 |
| ntd5 | 0.143 | ptmab | -0.092 |
| fgfrl1b | 0.136 | elavl3 | -0.089 |
| frem3 | 0.132 | marcksl1b | -0.087 |
| cx43 | 0.128 | h3f3d | -0.082 |
| si:ch211-66e2.5 | 0.128 | cirbpb | -0.080 |
| cyp2ad3 | 0.128 | hmgb3a | -0.078 |
| glula | 0.117 | myt1b | -0.076 |
| slc3a2a | 0.113 | tmsb | -0.075 |
| ppap2d | 0.113 | hnrnpaba | -0.075 |
| anxa4 | 0.112 | marcksb | -0.074 |
| cdo1 | 0.112 | tubb2b | -0.072 |
| pdgfrb | 0.110 | tuba1c | -0.072 |
| slc4a4a | 0.108 | si:dkey-276j7.1 | -0.071 |
| cyp3c1 | 0.108 | tubb5 | -0.071 |
| krt18a.1 | 0.105 | tmsb4x | -0.069 |
| aplnr2 | 0.103 | stmn1b | -0.067 |
| gdf6b | 0.101 | chd4a | -0.067 |
| cebpd | 0.100 | nova2 | -0.065 |
| fbn2b | 0.099 | sox11b | -0.063 |
| lamb1a | 0.098 | ptmaa | -0.062 |
| anxa13 | 0.098 | hsp90ab1 | -0.061 |
| sdc2 | 0.097 | elavl4 | -0.060 |
| emid1 | 0.096 | gng2 | -0.060 |
| spon2a | 0.095 | si:ch211-222l21.1 | -0.059 |
| sult6b1 | 0.095 | tmeff1b | -0.059 |
| apela | 0.094 | stx1b | -0.058 |
| anxa11b | 0.094 | zc4h2 | -0.058 |
| sparc | 0.094 | gng3 | -0.058 |
| mlc1 | 0.093 | nkain1 | -0.057 |
| igf2b | 0.093 | scrt2 | -0.057 |
| hepacama | 0.093 | mex3b | -0.057 |
| gstp1 | 0.091 | stxbp1a | -0.056 |
| gpc1b | 0.091 | si:ch211-288g17.3 | -0.056 |
| ptn | 0.091 | myt1a | -0.056 |