patched 2
ZFIN 
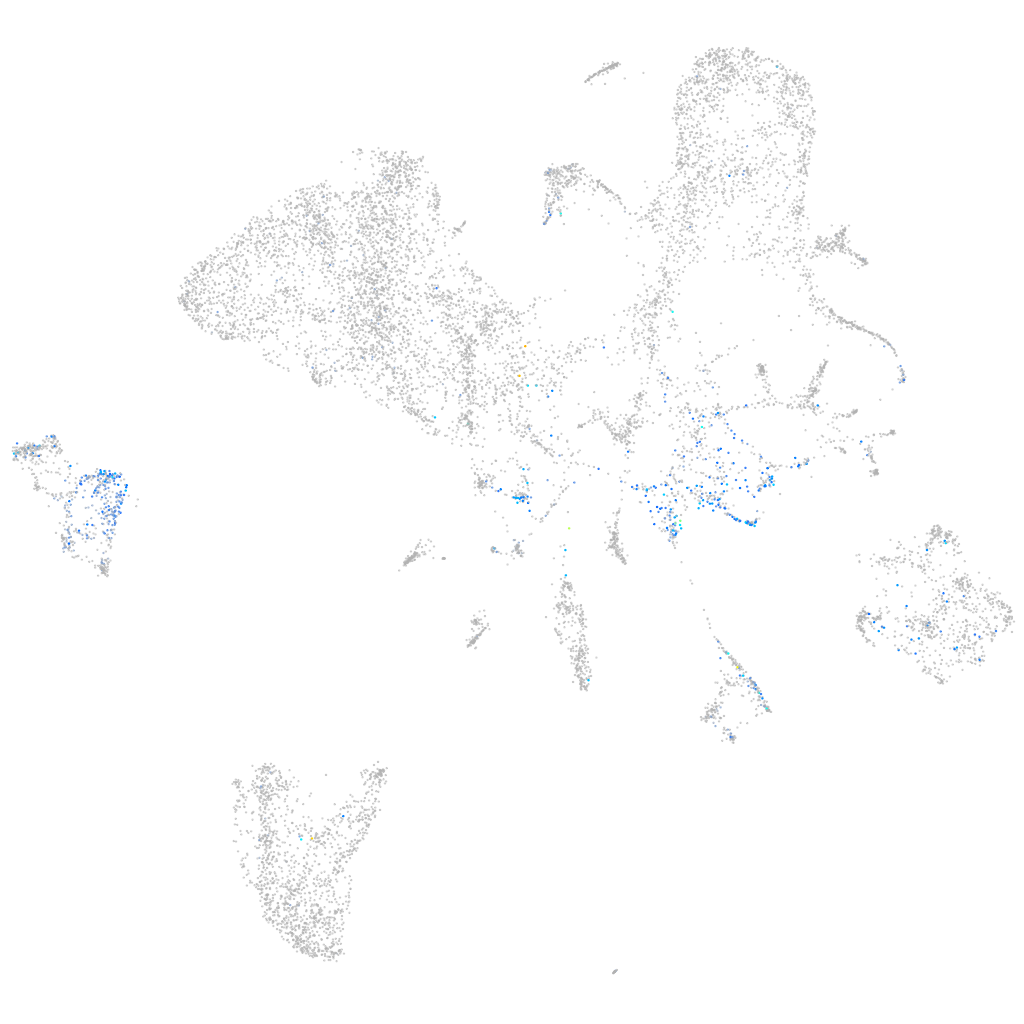
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bcam | 0.255 | gapdh | -0.210 |
| zfp36l1a | 0.254 | gamt | -0.174 |
| col2a1a | 0.248 | eno3 | -0.170 |
| hhip | 0.245 | pklr | -0.166 |
| ndnf | 0.242 | fbp1b | -0.166 |
| CABZ01075068.1 | 0.241 | gatm | -0.161 |
| fgfrl1b | 0.234 | gpx4a | -0.160 |
| pcdh19 | 0.234 | lgals2b | -0.158 |
| cd9b | 0.234 | ahcy | -0.153 |
| hoxb3a | 0.230 | apoa4b.1 | -0.152 |
| cldn7b | 0.228 | glud1b | -0.151 |
| si:rp71-77l1.1 | 0.228 | suclg1 | -0.148 |
| shhb | 0.227 | scp2a | -0.147 |
| si:busm1-71b9.3 | 0.224 | sod1 | -0.145 |
| fbn2b | 0.223 | apoc2 | -0.145 |
| nav3 | 0.223 | mat1a | -0.144 |
| fstl1b | 0.222 | cx32.3 | -0.142 |
| cldni | 0.221 | gstt1a | -0.142 |
| sox2 | 0.221 | BX908782.3 | -0.141 |
| si:dkey-261h17.1 | 0.219 | dap | -0.141 |
| txnipa | 0.218 | atp5mc1 | -0.139 |
| pitx1 | 0.218 | pgk1 | -0.139 |
| sparc | 0.217 | apoa1b | -0.137 |
| lin28a | 0.216 | abat | -0.135 |
| sall4 | 0.216 | agxta | -0.135 |
| col8a1a | 0.213 | gcshb | -0.135 |
| igfbp7 | 0.213 | sult2st2 | -0.134 |
| epcam | 0.213 | afp4 | -0.134 |
| cldn1 | 0.211 | tpi1b | -0.133 |
| fras1 | 0.210 | nipsnap3a | -0.133 |
| glulb | 0.209 | sod2 | -0.133 |
| col4a6 | 0.208 | bhmt | -0.132 |
| tpm4a | 0.207 | rdh1 | -0.132 |
| cldnb | 0.206 | pnp4b | -0.131 |
| sox11a | 0.204 | aldob | -0.130 |