proteasome activator subunit 2
ZFIN 
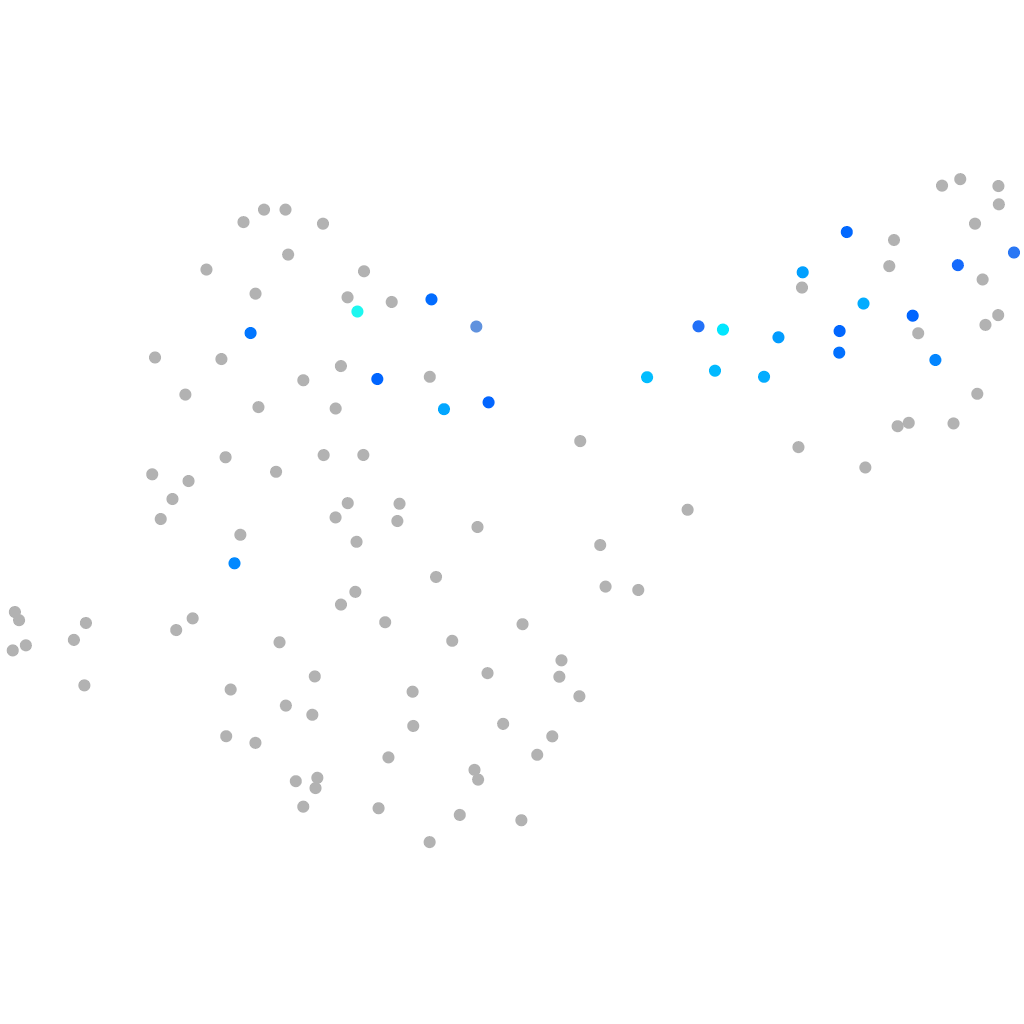
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| psmb8a | 0.784 | hnrnpaba | -0.640 |
| gchfr | 0.701 | ncl | -0.556 |
| pld6 | 0.687 | h1m | -0.521 |
| rab11bb | 0.676 | si:ch73-281n10.2 | -0.495 |
| fkbp1b | 0.672 | hnrnpa1b | -0.479 |
| zgc:63470 | 0.667 | ptmab | -0.479 |
| elovl2 | 0.667 | brd3a | -0.459 |
| tdrd12 | 0.662 | ccna2 | -0.459 |
| iars | 0.659 | akap12b | -0.456 |
| ldlrb | 0.657 | marcksb | -0.451 |
| b2m | 0.654 | hmgb1a | -0.446 |
| si:ch1073-314i13.4 | 0.653 | si:ch211-51e12.7 | -0.446 |
| crybg1a | 0.649 | stm | -0.444 |
| MCMDC2 | 0.648 | nucks1a | -0.435 |
| psmb12 | 0.645 | NC-002333.4 | -0.434 |
| dusp22b | 0.642 | ppig | -0.426 |
| cox5b2 | 0.635 | marcksl1b | -0.419 |
| sdhda | 0.634 | banf1 | -0.415 |
| si:ch211-262e15.1 | 0.629 | snrpa | -0.414 |
| phb2b | 0.628 | ddit4 | -0.412 |
| piwil1 | 0.626 | snrpb2 | -0.404 |
| psma6l | 0.625 | srrt | -0.404 |
| eno1b | 0.621 | anp32e | -0.397 |
| dele1 | 0.621 | ybx1 | -0.392 |
| sp100.2 | 0.620 | tbx16 | -0.386 |
| cox8b | 0.614 | tpx2 | -0.384 |
| psme1 | 0.614 | rbm4.3 | -0.382 |
| alcama | 0.606 | elavl1a | -0.373 |
| txndc5 | 0.606 | cd2bp2 | -0.369 |
| mcama | 0.606 | hnrnpa1a | -0.368 |
| mycbp | 0.606 | myl12.1 | -0.363 |
| fkbp6 | 0.603 | srrm1 | -0.360 |
| tcea2 | 0.601 | sde2 | -0.358 |
| hagh | 0.600 | syncrip | -0.358 |
| si:dkey-71b5.7 | 0.591 | hnrnpub | -0.355 |