"proteasome 26S subunit, non-ATPase 5"
ZFIN 
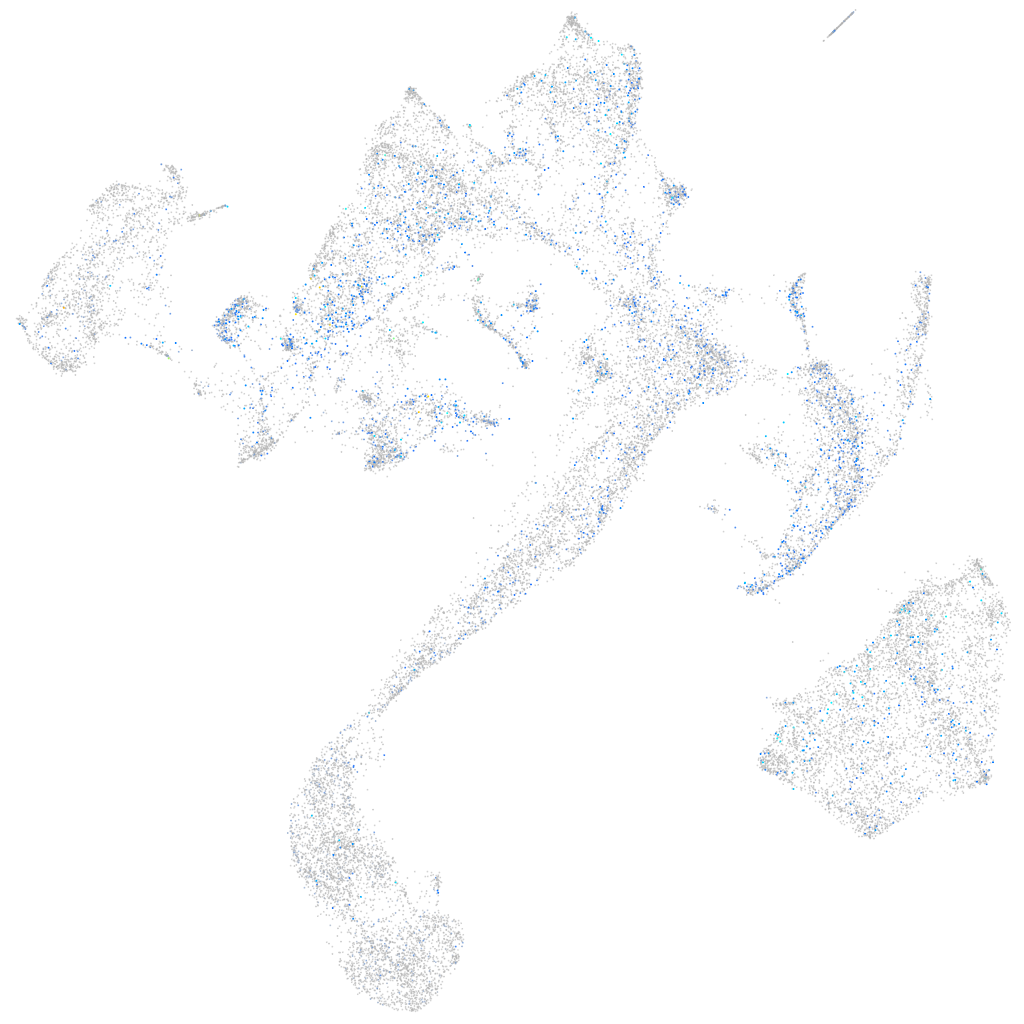
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpa0l | 0.113 | tnnc2 | -0.080 |
| h3f3a | 0.113 | ldb3a | -0.077 |
| dut | 0.113 | gapdh | -0.076 |
| ppiab | 0.106 | actn3a | -0.076 |
| cfl1 | 0.104 | acta1b | -0.076 |
| zgc:56493 | 0.104 | tpma | -0.075 |
| hmgn2 | 0.104 | neb | -0.074 |
| prdx2 | 0.101 | actn3b | -0.074 |
| psma3 | 0.098 | si:ch73-367p23.2 | -0.073 |
| psmb7 | 0.096 | ak1 | -0.073 |
| tubb2b | 0.095 | nme2b.2 | -0.072 |
| psma5 | 0.095 | myom1a | -0.071 |
| snrpf | 0.094 | ttn.1 | -0.071 |
| ewsr1b | 0.093 | ldb3b | -0.071 |
| psmb1 | 0.092 | aldoab | -0.071 |
| hmgb2b | 0.092 | ckmb | -0.070 |
| h2afvb | 0.092 | smyd1a | -0.069 |
| mif | 0.092 | actc1b | -0.069 |
| alyref | 0.092 | XLOC-025819 | -0.069 |
| ran | 0.092 | XLOC-005350 | -0.069 |
| tubb4b | 0.090 | atp2a1 | -0.069 |
| lsm7 | 0.090 | srl | -0.068 |
| actb1 | 0.090 | si:ch211-266g18.10 | -0.068 |
| mcm7 | 0.089 | pgam2 | -0.068 |
| tuba8l4 | 0.089 | XLOC-001975 | -0.068 |
| sub1a | 0.088 | hhatla | -0.068 |
| tuba8l3 | 0.088 | tmod4 | -0.067 |
| psmb6 | 0.088 | mybphb | -0.067 |
| psma6a | 0.088 | ttn.2 | -0.067 |
| her15.1 | 0.088 | casq2 | -0.067 |
| cct2 | 0.087 | ckma | -0.066 |
| eif3ba | 0.087 | mylz3 | -0.066 |
| h2afva | 0.086 | XLOC-006515 | -0.066 |
| calm3b | 0.086 | rtn2a | -0.066 |
| snrpd2 | 0.085 | jph2 | -0.066 |