proteasome 20S subunit beta 8A
ZFIN 
Other cell groups


all cells |
blastomeres |
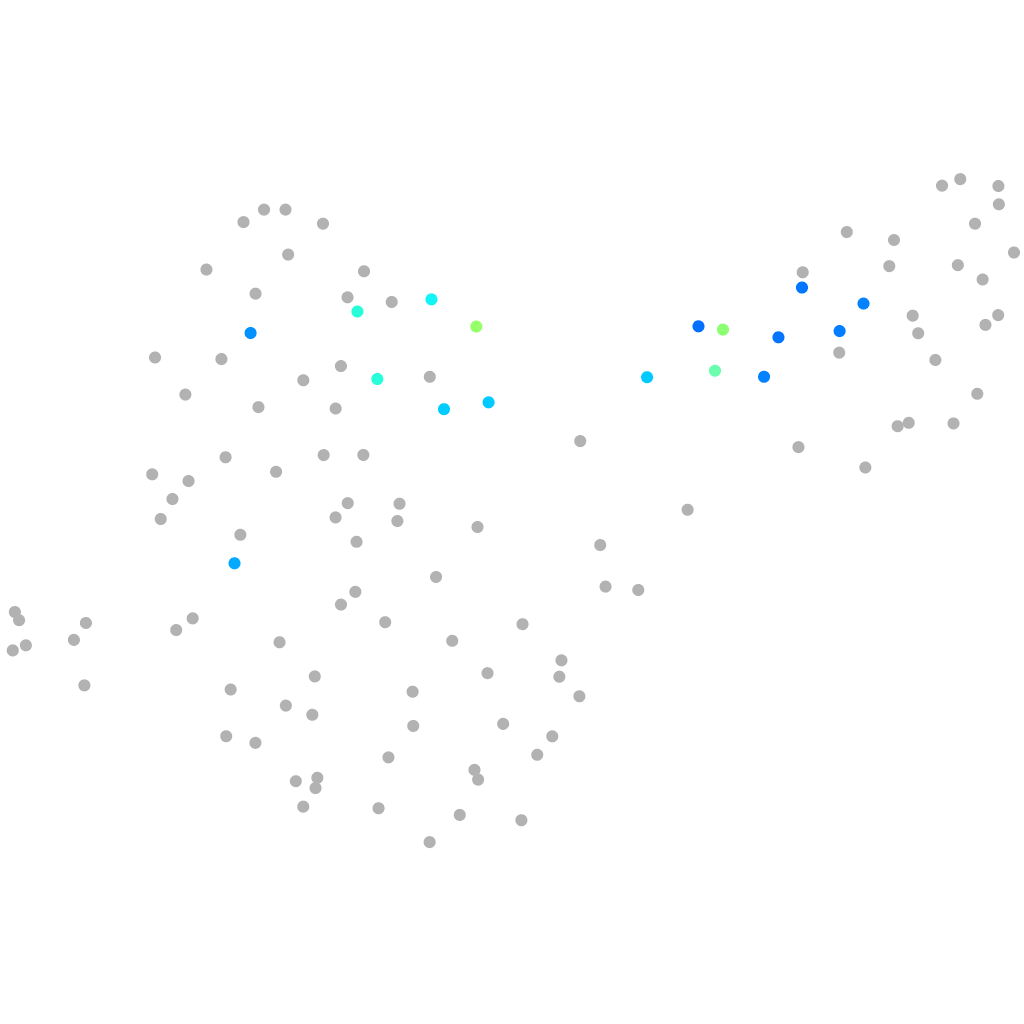
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| piwil1 | 0.794 | hnrnpaba | -0.637 |
| psme2 | 0.784 | h1m | -0.524 |
| cox5b2 | 0.758 | actb1 | -0.519 |
| alcama | 0.753 | hmga1a | -0.502 |
| LOC101885930 | 0.734 | ncl | -0.470 |
| atr | 0.722 | ccna2 | -0.456 |
| isg15 | 0.710 | ptmab | -0.439 |
| sdhda | 0.693 | anp32e | -0.436 |
| exd1 | 0.688 | hmgb2a | -0.429 |
| fkbp6 | 0.680 | si:ch73-281n10.2 | -0.428 |
| gchfr | 0.675 | akap12b | -0.428 |
| ifi45 | 0.674 | hnrnpa1b | -0.421 |
| b2m | 0.674 | stm | -0.415 |
| hormad1 | 0.672 | nucks1a | -0.412 |
| piwil2 | 0.670 | syncrip | -0.412 |
| tuba7l | 0.667 | ctsla | -0.411 |
| CU571163.1 | 0.663 | anp32b | -0.405 |
| pld6 | 0.661 | snrpa | -0.403 |
| stag3 | 0.659 | hspb1 | -0.401 |
| mycbp | 0.658 | hmgb1a | -0.390 |
| tdrd9 | 0.651 | hnrnpub | -0.383 |
| cox8b | 0.650 | ybx1 | -0.382 |
| psmb9a | 0.646 | NC-002333.4 | -0.379 |
| sp100.2 | 0.644 | epcam | -0.376 |
| tdrd12 | 0.641 | cx43.4 | -0.375 |
| iars | 0.636 | ppig | -0.375 |
| psmb12 | 0.634 | brd3a | -0.371 |
| LOC100149647 | 0.633 | cd2bp2 | -0.370 |
| dele1 | 0.630 | marcksb | -0.368 |
| hnrnpa3 | 0.629 | srrt | -0.368 |
| fam102ba | 0.628 | marcksl1b | -0.366 |
| si:ch211-214c7.5 | 0.627 | dnajc1 | -0.363 |
| hyi | 0.626 | rbm7 | -0.360 |
| plppr5b | 0.621 | myl12.1 | -0.360 |
| irak4 | 0.621 | ddit4 | -0.359 |